Comparative Methods for Molecular Determination of Host-Specificity Factors in Plant-Pathogenic Fungi
Abstract
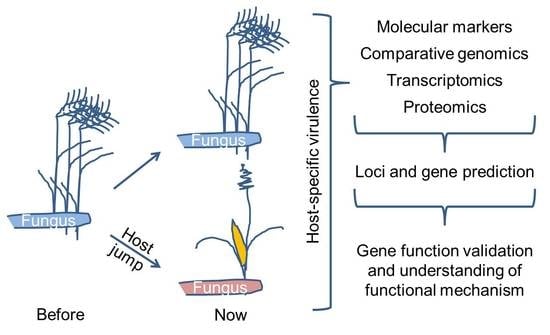
:1. Introduction
2. Defining and Characterizing the Pathosystem
3. Using Molecular Markers for Genotyping
4. Comparing Whole Genome Sequences
5. Comparing Complete Proteomes
6. Investigating Segregating Populations
7. Validating Functional Contribution to Host-Specificity
8. Deciphering Functional Mechanisms of Host-Specificity
9. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Van Der Biezen, E.A.; Jones, J.D.G. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 1998, 23, 454–456. [Google Scholar] [CrossRef]
- Van der Hoorn, R.A.L.; Kamoun, S. From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 2008, 20, 2009–2017. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- De Jonge, R.; Bolton, M.D.; Thomma, B.P.H.J. How filamentous pathogens co-opt plants: The ins and outs of fungal effectors. Curr. Opin. Plant Biol. 2011, 14, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Petit-Houdenot, Y.; Fudal, I. Complex interactions between fungal avirulence genes and their corresponding plant resistance genes and consequences for disease resistance management. Front. Plant Sci. 2017, 8, 1072. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Vy, T.T.P.; Yoshida, K.; Asano, H.; Mitsuoka, C.; Asuke, S.; Anh, V.L.; Cumagun, C.J.R.; Chuma, I.; Terauchi, R.; et al. Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science 2017, 357, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Meena, M.; Gupta, S.K.; Swapnil, P.; Zehra, A.; Dubey, M.K.; Upadhyay, R.S. Alternaria toxins: Potential virulence factors and genes related to pathogenesis. Front. Microbiol. 2017, 8, 1451. [Google Scholar] [CrossRef] [PubMed]
- Hatta, R.; Ito, K.; Hosaki, Y.; Tanaka, T.; Tanaka, A.; Yamamoto, M.; Akimitsu, K.; Tsuge, T. A conditionally dispensable chromosome controls host-specific pathogenicity in the fungal plant pathogen Alternaria alternata. Genetics 2002, 161, 59–70. [Google Scholar] [PubMed]
- Tsuge, T.; Harimoto, Y.; Hanada, K.; Akagi, Y.; Kodama, M.; Akimitsu, K.; Yamamoto, M. Evolution of pathogenicity controlled by small, dispensable chromosomes in Alternaria alternata pathogens. Physiol. Mol. Plant Pathol. 2016, 95, 27–31. [Google Scholar] [CrossRef]
- Akagi, Y.; Akamatsu, H.; Otani, H.; Kodama, M. Horizontal chromosome transfer, a mechanism for the evolution and differentiation of a plant-pathogenic fungus. Eukaryot. Cell 2009, 8, 1732–1738. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, G.; Person, C. Genetic control of virulence in Ustilago hordei III. Identification of genes for host resistance and demonstration of gene-for-gene relations. Can. J. Genet. Cytol. 1972, 14, 209–213. [Google Scholar] [CrossRef]
- Linning, R.; Lin, D.; Lee, N.; Abdennadher, M.; Gaudet, D.; Thomas, P.; Mills, D.; Kronstad, J.W.; Bakkeren, G. Marker-based cloning of the region containing the Uhavr1 avirulence gene from the basidiomycete barley pathogen Ustilago hordei. Genetics 2004, 166, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Laurie, J.D.; Linning, R.; Cervantes-Chavez, J.A.; Gaudet, D.; Bakkeren, G. An immunity-triggering effector from the barley smut fungus Ustilago hordei resides in an Ustilaginaceae-specific cluster bearing signs of transposable element-assisted evolution. PLoS Pathog. 2014, 10, e1004223. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Kemppainen, M.; Kale, S.D.; Kohler, A.; Legue, V.; Brun, A.; Tyler, B.M.; Pardo, A.G.; Martin, F. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr. Biol. 2011, 21, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Van der Does, H.C.; Rep, M. Virulence genes and the evolution of host specificity in plant-pathogenic fungi. Mol. Plant Microbe Interact. 2007, 20, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Mehrabi, R.; Bahkali, A.H.; Abd-Elsalam, K.A.; Moslem, M.; M’barek, S.B.; Gohari, A.M.; Jashni, M.K.; Stergiopoulos, I.; Kema, G.H.; de Wit, P.J. Horizontal gene and chromosome transfer in plant pathogenic fungi affecting host range. FEMS Microbiol. Rev. 2011, 35, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Andrie, R.M.; Pandelova, I.; Ciuffetti, L.M. A combination of phenotypic and genotypic characterization strengthens Pyrenophora tritici-repentis race identification. Phytopathology 2007, 97, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Chiapello, H.; Mallet, L.; Guerin, C.; Aguileta, G.; Amselem, J.; Kroj, T.; Ortega-Abboud, E.; Lebrun, M.H.; Henrissat, B.; Gendrault, A.; et al. Deciphering genome content and evolutionary relationships of isolates from the fungus Magnaporthe oryzae attacking different host plants. Genome Biol. Evol. 2015, 7, 2896–2912. [Google Scholar] [CrossRef] [PubMed]
- Baayen, R.P.; O’Donnell, K.; Bonants, P.J.M.; Cigelnik, E.; Kroon, L.P.N.M.; Roebroeck, E.J.A.; Waalwijk, C. Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology 2000, 90, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-H.; Sheu, Z.-M.; Wang, T.-C. Host specificity and tomato-related race composition of Phytophthora infestans isolates in Taiwan during 2004 and 2005. Plant Dis. 2008, 92, 751–755. [Google Scholar] [CrossRef]
- Kovacikova, E.; Kratka, J. Different manifestations of the pathogenity of some strains of Fusarium oxysporum f. sp. pisi. Zentralbl. Bakteriol. Naturwiss. 1979, 134, 159–166. [Google Scholar] [PubMed]
- Heath, M.C. Host species specificity of the goldenrod rust fungus and the existence of rust resistance within some Goldenrod species. Can. J. Bot. 1992, 70, 2461–2466. [Google Scholar] [CrossRef]
- Bernstein, B.; Zehr, E.I.; Dean, R.A.; Shabi, E. Characteristics of colletotrichum from peach, apple, pecan, and other hosts. Plant Dis. 1995, 79, 478–482. [Google Scholar] [CrossRef]
- Orlicz-Luthardt, A.; Rieckmann, U.; Dercks, W. Pathogenicity of some species of Fusarium and formae speciales to china aster and carnation. Gartenbauwissenschaft 2000, 65, 137–143. [Google Scholar]
- Peever, T.L.; Olsen, L.; Ibanez, A.; Timmer, L.W. Genetic differentiation and host specificity among populations of Alternaria spp. causing brown spot of grapefruit and tangerine × grapefruit hybrids in Florida. Phytopathology 2000, 90, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Yehuda, P.B.; Eilam, T.; Manisterski, J.; Shimoni, A.; Anikster, Y. Leaf rust on Aegilops speltoides caused by a new forma specialis of Puccinia triticina. Phytopathology 2004, 94, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Bello, M.A.; Chilvers, M.I.; Akamatsu, H.; Peever, T.L. Host specificity of Ascochyta spp. infecting legumes of the Viciae and Cicerae tribes and pathogenicity of an interspecific hybrid. Phytopathology 2006, 96, 1148–1156. [Google Scholar] [CrossRef] [PubMed]
- Llorens, A.; Hinojo, M.J.; Mateo, R.; Gonzalez-Jaen, M.T.; Valle-Algarra, F.M.; Logrieco, A.; Jimenez, M. Characterization of Fusarium spp. isolates by PCR-RFLP analysis of the intergenic spacer region of the rRNA gene (rDNA). Int. J. Food Microbiol. 2006, 106, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Hedh, J.; Johansson, T.; Tunlid, A. Variation in host specificity and gene content in strains from genetically isolated lineages of the ectomycorrhizal fungus Paxillus involutus s. lat. Mycorrhiza 2009, 19, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Greco, M.; Patriarca, A.; Terminiello, L.; Fernandez Pinto, V.; Pose, G. Toxigenic Alternaria species from Argentinean blueberries. Int. J. Food Microbiol. 2012, 154, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, B.E.; Pringle, A. Geographically structured host specificity is caused by the range expansions and host shifts of a symbiotic fungus. ISME J. 2012, 6, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Zuther, K.; Kahnt, J.; Utermark, J.; Imkampe, J.; Uhse, S.; Schirawski, J. Host specificity of Sporisorium reilianum is tightly linked to generation of the phytoalexin luteolinidin by Sorghum bicolor. Mol. Plant Microbe Interact. 2012, 25, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Beckstead, J.; Meyer, S.E.; Ishizuka, T.S.; McEvoy, K.M.; Coleman, C.E. Lack of host specialization on winter annual grasses in the fungal seed bank pathogen Pyrenophora semeniperda. PLoS ONE 2016, 11, e0151058. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, K.; Kojima, M. The role of stress metabolites in establishing host-parasite specificity between sweet potato and Ceratocystis fimbriata, black rot fungus. Agric. Biol. Chem. 1986, 50, 1839–1846. [Google Scholar] [CrossRef]
- Masunaka, A.; Ohtani, K.; Peever, T.L.; Timmer, L.W.; Tsuge, T.; Yamamoto, M.; Yamamoto, H.; Akimitsu, K. An isolate of Alternaria alternata that is pathogenic to both tangerines and rough lemon and produces two host-selective toxins, ACT- and ACR-toxins. Phytopathology 2005, 95, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Kema, G.H.J.; Waalwijk, C.; McDonald, B.A. Distribution of mating type alleles in the wheat pathogen Mycosphaerella graminicola over spatial scales from lesions to continents. Fungal Genet. Biol. 2002, 36, 128–136. [Google Scholar] [CrossRef]
- Sone, T.; Suto, M.; Tomita, F. Host species-specific repetitive DNA sequence in the genome of Magnaporthe grisea, the rice blast fungus. Biosci. Biotechnol. Biochem. 1993, 57, 1228–1230. [Google Scholar] [CrossRef] [PubMed]
- Maharaj, A.; Rampersad, S.N. Genetic differentiation of Colletotrichum gloeosporioides and C. truncatum associated with anthracnose disease of papaya (Carica papaya L.) and bell pepper (Capsium annuum L.) based on ITS PCR-RFLP fingerprinting. Mol. Biotechnol. 2012, 50, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Levis, C.; Giraud, T.; Dutertre, M.; Fortini, D.; Brygoo, Y. Telomeric DNA of Botrytis cinerea: A useful tool for strain identification. FEMS Microbiol. Lett. 1997, 157, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Viji, G.; Wu, B.; Kang, S.; Uddin, W.; Huff, D.R. Pyricularia grisea causing gray leaf spot of perennial ryegrass turf: Population structure and host specificity. Plant Dis. 2001, 85, 817–826. [Google Scholar] [CrossRef]
- Quecine, M.C.; Bini, A.P.; Romagnoli, E.R.; Andreote, F.D.; Moon, D.H.; Labate, C.A. Genetic variability in Puccinia psidii populations as revealed by PCR-DGGE and T-RFLP markers. Plant Dis. 2014, 98, 16–23. [Google Scholar] [CrossRef]
- Zaffarano, P.L.; McDonald, B.A.; Linde, C.C. Two new species of Rhynchosporium. Mycologia 2011, 103, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Attitalla, I.H.; Fatehi, J.; Levenfors, J.; Brishammar, S. A rapid molecular method for differentiating two special forms (lycopersici and radicis-lycopersici) of Fusarium oxysporum. Mycol. Res. 2004, 108, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Nechwatal, J.; Wielgoss, A.; Mendgen, K. Diversity, host, and habitat specificity of oomycete communities in declining reed stands (Phragmites australis) of a large freshwater lake. Mycol. Res. 2008, 112, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Oyarzun, P.J.; Pozo, A.; Ordonez, M.E.; Doucett, K.; Forbes, G.A. Host Specificity of Phytophthora infestans on tomato and potato in Ecuador. Phytopathology 1998, 88, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Garry, G.; Forbes, G.A.; Salas, A.; Santa Cruz, M.; Perez, W.G.; Nelson, R.J. Genetic diversity and host differentiation among isolates of Phytophthora infestans from cultivated potato and wild solanaceous hosts in Peru. Plant Pathol. 2005, 54, 740–748. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Douhan, G.W.; de la Cerda, K.A.; Huryn, K.L.; Greer, C.A.; Wong, F.P. Contrasting genetic structure between Magnaporthe grisea populations associated with the golf course turfgrasses Lolium perenne (perennial ryegrass) and Pennisetum clandestinum (kikuyugrass). Phytopathology 2011, 101, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Flier, W.G.; Grünwald, N.J.; Kroon, L.P.N.M.; Sturbaum, A.K.; van den Bosch, T.B.M.; Garay-Serrano, E.; Lozoya-Saldaña, H.; Fry, W.E.; Turkensteen, L.J. The population structure of Phytophthora infestans from the toluca valley of central Mexico suggests genetic differentiation between populations from cultivated potato and wild Solanum spp. Phytopathology 2003, 93, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Karimi, S.; Mirlohi, A.; Sabzalian, M.R.; Sayed Tabatabaei, B.E.; Sharifnabi, B. Molecular evidence for Neotyphodium fungal endophyte variation and specificity within host grass species. Mycologia 2012, 104, 1281–1290. [Google Scholar] [CrossRef] [PubMed]
- Wyand, R.A.; Brown, J.K. Genetic and forma specialis diversity in Blumeria graminis of cereals and its implications for host-pathogen co-evolution. Mol. Plant Pathol. 2003, 4, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wasilwa, L.A.; Morelock, T.E.; O’Neill, N.R.; Correll, J.C. Comparison of Colletotrichum orbiculare and several allied Colletotrichum spp. for mtDNA RFLPs, intron RFLP and sequence variation, vegetative compatibility, and host specificity. Phytopathology 2007, 97, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Rigotti, S.; Gindro, K.; Richter, H.; Viret, O. Characterization of molecular markers for specific and sensitive detection of Botrytis cinerea Pers.: Fr. in strawberry (Fragariaxananassa duch.) using PCR. FEMS Microbiol. Lett. 2002, 209, 169–174. [Google Scholar] [CrossRef]
- Mahmodi, F.; Kadir, J.B.; Puteh, A.; Pourdad, S.S.; Nasehi, A.; Soleimani, N. Genetic diversity and differentiation of Colletotrichum spp. isolates associated with Leguminosae using multigene loci, RAPD and ISSR. Plant Pathol. 2014, 30, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Peever, T.L.; Canihos, Y.; Olsen, L.; Ibanez, A.; Liu, Y.C.; Timmer, L.W. Population genetic structure and host specificity of Alternaria spp. causing brown spot of minneola tangelo and rough lemon in Florida. Phytopathology 1999, 89, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Lievens, B.; Claes, L.; Vakalounakis, D.J.; Vanachter, A.C.; Thomma, B.P. A robust identification and detection assay to discriminate the cucumber pathogens Fusarium oxysporum f. sp. cucumerinum and f. sp. radicis-cucumerinum. Environ. Microbiol. 2007, 9, 2145–2161. [Google Scholar]
- King, K.M.; West, J.S.; Brunner, P.C.; Dyer, P.S.; Fitt, B.D. Evolutionary relationships between Rhynchosporium lolii sp. nov. and other Rhynchosporium species on grasses. PLoS ONE 2013, 8, e72536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Chao, G.M.; Liu, J.J.; Zhu, M.Q.; Wang, Y.; Feng, B.L. Genetic diversity of Ustilago hordei in Tibetan areas as revealed by RAPD and SSR. J. Integr. Agric. 2016, 15, 2299–2308. [Google Scholar] [CrossRef]
- Perlin, M.H.; Hughes, C.; Welch, J.; Akkaraju, S.; Steinecker, D.; Kumar, A.; Smith, B.; Garr, S.S.; Brown, S.A.; Andom, T. Molecular approaches to differentiate subpopulations or formae speciales of the fungal phytopathogen Microbotryum violaceum. Int. J. Plant Sci. 1997, 158, 568–574. [Google Scholar] [CrossRef]
- Santini, A.; Capretti, P. Analysis of the Italian population of Ceratocystis fimbriata f.sp. platani using RAPD and minisatellite markers. Plant Pathol. 2000, 49, 461–467. [Google Scholar]
- Freeman, S.; Katan, T.; Shabi, E. Characterization of Colletotrichum gloeosporioides isolates from avocado and almond fruits with molecular and pathogenicity tests. Appl. Environ. Microbiol. 1996, 62, 1014–1020. [Google Scholar] [PubMed]
- Asadollahi, M.; Fekete, E.; Karaffa, L.; Flipphi, M.; Arnyasi, M.; Esmaeili, M.; Vaczy, K.Z.; Sandor, E. Comparison of Botrytis cinerea populations isolated from two open-field cultivated host plants. Microbiol. Res. 2013, 168, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Van Putten, W.F.; Biere, A.; Van Damme, J.M.M. Host-related genetic differentiation in the anther smut fungus Microbotryum violaceum in sympatric, parapatric and allopatric populations of two host species Silene latifolia and S. dioica. J. Evol. Biol. 2005, 18, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Dipietro, A.; Anaya, N.; Roncero, M.I.G. Occurrence of a retrotransposon-like sequence among different formae speciales and races of Fusarium oxysporum. Mycol. Res. 1994, 98, 993–996. [Google Scholar] [CrossRef]
- Samuel, S.; Veloukas, T.; Papavasileiou, A.; Karaoglanidis, G.S. Differences in frequency of transposable elements presence in Botrytis cinerea populations from several hosts in Greece. Plant Dis. 2012, 96, 1286–1290. [Google Scholar] [CrossRef]
- Laurie, J.D.; Ali, S.; Linning, R.; Mannhaupt, G.; Wong, P.; Guldener, U.; Munsterkotter, M.; Moore, R.; Kahmann, R.; Bakkeren, G.; et al. Genome comparison of barley and maize smut fungi reveals targeted loss of RNA silencing components and species-specific presence of transposable elements. Plant Cell 2012, 24, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Schirawski, J.; Mannhaupt, G.; Munch, K.; Brefort, T.; Schipper, K.; Doehlemann, G.; Di Stasio, M.; Rossel, N.; Mendoza-Mendoza, A.; Pester, D.; et al. Pathogenicity determinants in smut fungi revealed by genome comparison. Science 2010, 330, 1546–1548. [Google Scholar] [CrossRef] [PubMed]
- Dutheil, J.Y.; Mannhaupt, G.; Schweizer, G.; Sieber, C.M.K.; Munsterkotter, M.; Guldener, U.; Schirawski, J.; Kahmann, R. A tale of genome compartmentalization: The evolution of virulence lusters in smut fungi. Genome Biol. Evol. 2016, 8, 681–704. [Google Scholar] [CrossRef] [PubMed]
- Buiate, E.A.; Xavier, K.V.; Moore, N.; Torres, M.F.; Farman, M.L.; Schardl, C.L.; Vaillancourt, L.J. A comparative genomic analysis of putative pathogenicity genes in the host-specific sibling species Colletotrichum graminicola and Colletotrichum sublineola. BMC Genom. 2017, 18, 67. [Google Scholar] [CrossRef] [PubMed]
- Penselin, D.; Munsterkotter, M.; Kirsten, S.; Felder, M.; Taudien, S.; Platzer, M.; Ashelford, K.; Paskiewicz, K.H.; Harrison, R.J.; Hughes, D.J.; et al. Comparative genomics to explore phylogenetic relationship, cryptic sexual potential and host specificity of Rhynchosporium species on grasses. BMC Genom. 2016, 17, 953. [Google Scholar] [CrossRef] [PubMed]
- Niehaus, E.M.; Munsterkotter, M.; Proctor, R.H.; Brown, D.W.; Sharon, A.; Idan, Y.; Oren-Young, L.; Sieber, C.M.; Novak, O.; Pencik, A.; et al. Comparative “Omics” of the Fusarium fujikuroi species complex highlights differences in genetic potential and metabolite synthesis. Genome Biol. Evol. 2016, 8, 3574–3599. [Google Scholar] [CrossRef] [PubMed]
- Baroncelli, R.; Amby, D.B.; Zapparata, A.; Sarrocco, S.; Vannacci, G.; Le Floch, G.; Harrison, R.J.; Holub, E.; Sukno, S.A.; Sreenivasaprasad, S.; et al. Gene family expansions and contractions are associated with host range in plant pathogens of the genus Colletotrichum. BMC Genom. 2016, 17, 555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, R.; Mishra, B.; Runge, F.; Thines, M. Gene loss rather than gene gain is associated with a host jump from monocots to dicots in the smut fungus Melanopsichium pennsylvanicum. Genome Biol. Evol. 2014, 6, 2034–2049. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.Y.; Zhao, W.S.; Chen, Z.; Xing, Q.K.; Zhang, W.; Chethana, K.W.T.; Xue, M.F.; Xu, J.P.; Phillips, A.J.L.; Wang, Y.; et al. Comparative genome and transcriptome analyses reveal adaptations to opportunistic infections in woody plant degrading pathogens of Botryosphaeriaceae. DNA Res. 2018, 25, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-J.; van der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.-J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.H.; Sharma, M.; Thatcher, L.F.; Azam, S.; Hane, J.K.; Sperschneider, J.; Kidd, B.N.; Anderson, J.P.; Ghosh, R.; Garg, G.; et al. Comparative genomics and prediction of conditionally dispensable sequences in legume-infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genom. 2016, 17, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Dam, P.; Fokkens, L.; Schmidt, S.M.; Linmans, J.H.; Kistler, H.C.; Ma, L.J.; Rep, M. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 2016, 18, 4087–4102. [Google Scholar] [CrossRef] [PubMed]
- Van der Does, H.C.; Lievens, B.; Claes, L.; Houterman, P.M.; Cornelissen, B.J.; Rep, M. The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ. Microbiol. 2008, 10, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
- Lievens, B.; Houterman, P.M.; Rep, M. Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 2009, 300, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Bhadauria, V.; MacLachlan, R.; Pozniak, C.; Banniza, S. Candidate effectors contribute to race differentiation and virulence of the lentil anthracnose pathogen Colletotrichum lentis. BMC Genom. 2015, 16, 628. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Ji, C.; Li, Y.; Wang, Z. Comparative transcriptomic analysis of race 1 and race 4 of Fusarium oxysporum f. sp. cubense induced with different carbon sources. G3 (Bethesda) 2017, 7, 2125–2138. [Google Scholar] [CrossRef] [PubMed]
- Kema, G.H.J.; Gohari, A.M.; Aouini, L.; Gibriel, H.A.Y.; Ware, S.B.; van den Bosch, F.; Manning-Smith, R.; Alonso-Chavez, V.; Helps, J.; Ben, M’ Barek, S.; et al. Stress and sexual reproduction affect the dynamics of the wheat pathogen effector AvrStb6 and strobilurin resistance. Nat. Genet. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.; Resjo, S.; Liu, Y.; Durling, M.; Heyman, F.; Levander, F.; Liu, Y.; Elfstrand, M.; Funck Jensen, D.; Andreasson, E.; et al. Comparative proteomic analysis of hyphae and germinating cysts of Phytophthora pisi and Phytophthora sojae. J. Proteom. 2015, 117, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Savidor, A.; Donahoo, R.S.; Hurtado-Gonzales, O.; Land, M.L.; Shah, M.B.; Lamour, K.H.; McDonald, W.H. Cross-species global proteomics reveals conserved and unique processes in Phytophthora sojae and Phytophthora ramorum. Mol. Cell. Proteom. 2008, 7, 1501–1516. [Google Scholar] [CrossRef] [PubMed]
- Quecine, M.C.; Leite, T.F.; Bini, A.P.; Regiani, T.; Franceschini, L.M.; Budzinski, I.G.F.; Marques, F.G.; Labate, M.T.V.; Guidetti-Gonzalez, S.; Moon, D.H.; et al. Label-free quantitative proteomic analysis of Puccinia psidii uredospores reveals differences of fungal populations infecting eucalyptus and guava. PLoS ONE 2016, 11, e0145343. [Google Scholar] [CrossRef] [PubMed]
- Bregar, O.; Mandelc, S.; Celar, F.; Javornik, B. Proteome analysis of the plant pathogenic fungus Monilinia laxa showing host specificity. Food Technol. Biotechnol. 2012, 50, 326–333. [Google Scholar]
- Cisar, C.R.; Spiegel, F.W.; TeBeest, D.O.; Trout, C. Evidence for mating between isolates of Colletotrichum gloeosporioides with different host specificities. Curr. Genet. 1994, 25, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Cozijnsen, A.J.; Popa, K.M.; Purwantara, A.; Rolls, B.D.; Howlett, B.J. Genome analysis of the plant pathogenic ascomycete Leptosphaeria maculans; mapping mating type and host specificity loci. Mol. Plant Pathol. 2000, 1, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Tosa, Y.; Tamba, H.; Tanaka, K.; Mayama, S. Genetic analysis of host species specificity of Magnaporthe oryzae isolates from rice and wheat. Phytopathology 2006, 96, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Murakami, J.; Tomita, R.; Kataoka, T.; Nakayashiki, H.; Tosa, Y.; Mayama, S. Analysis of host species specificity of Magnaporthe grisea toward foxtail millet using a genetic cross between isolates from wheat and foxtail millet. Phytopathology 2003, 93, 42–45. [Google Scholar] [CrossRef] [PubMed]
- Foulongne-Oriol, M. Genetic linkage mapping in fungi: Current state, applications, and future trends. Appl. Microbiol. Biotechnol. 2012, 95, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Poloni, A.; Schirawski, J. Host specificity in Sporisorium reilianum is determined by distinct mechanisms in maize and sorghum. Mol. Plant Pathol. 2016, 17, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Valent, B.; Farrall, L.; Chumley, F.G. Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 1991, 127, 87–101. [Google Scholar] [PubMed]
- Kang, S.; Sweigard, J.A.; Valent, B. The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 1995, 8, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Sweigard, J.A.; Carroll, A.M.; Kang, S.; Farrall, L.; Chumley, F.G.; Valent, B. Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 1995, 7, 1221–1233. [Google Scholar] [CrossRef] [PubMed]
- Peyyala, R.; Farman, M.L. Magnaporthe oryzae isolates causing gray leaf spot of perennial ryegrass possess a functional copy of the AVR1-CO39 avirulence gene. Mol. Plant Pathol. 2006, 7, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Meijer, M.; Houterman, P.M.; van der Does, H.C.; Cornelissen, B.J. Fusarium oxysporum evades I-3-mediated resistance without altering the matching avirulence gene. Mol. Plant Microbe Interact. 2005, 18, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; van der Does, H.C.; Meijer, M.; van Wijk, R.; Houterman, P.M.; Dekker, H.L.; de Koster, C.G.; Cornelissen, B.J. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 2004, 53, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Wang, G.; Xiao, J.; Ling, J.; Yang, Y.; Xie, B. A SIX1 homolog in Fusarium oxysporum f. sp. conglutinans is required for full virulence on cabbage. PLoS ONE 2016, 11, e0152273. [Google Scholar]
- Poppe, S.; Dorsheimer, L.; Happel, P.; Stukenbrock, E.H. Rapidly evolving genes are key players in host specialization and virulence of the fungal wheat pathogen Zymoseptoria tritici (Mycosphaerella graminicola). PLOS Pathog. 2015, 11, e1005055. [Google Scholar] [CrossRef] [PubMed]
- Isshiki, A.; Akimitsu, K.; Yamamoto, M.; Yamamoto, H. Endopolygalacturonase is essential for citrus black rot caused by Alternaria citri but not brown spot caused by Alternaria alternata. Mol. Plant Microbe Interact. 2001, 14, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Andrie, R.M.; Ciuffetti, L.M. Pyrenophora bromi, causal agent of brownspot of bromegrass, expresses a gene encoding a protein with homology and similar activity to Ptr ToxB, a host-selective toxin of wheat. Mol. Plant Microbe Interact. 2011, 24, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Birch, P.R.; Armstrong, M.; Bos, J.; Boevink, P.; Gilroy, E.M.; Taylor, R.M.; Wawra, S.; Pritchard, L.; Conti, L.; Ewan, R.; et al. Towards understanding the virulence functions of RXLR effectors of the oomycete plant pathogen Phytophthora infestans. J. Exp. Bot. 2009, 60, 1133–1140. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.M.; Stam, R.; Cano, L.M.; Song, J.; Sklenar, J.; Yoshida, K.; Bozkurt, T.O.; Oliva, R.; Liu, Z.Y.; Tian, M.Y.; et al. Effector specialization in a lineage of the Irish potato famine pathogen. Science 2014, 343, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, K.; Yamamoto, H.; Akimitsu, K. Sensitivity to Alternaria alternata toxin in citrus because of altered mitochondrial RNA processing. Proc. Natl. Acad. Sci. USA 2002, 99, 2439–2444. [Google Scholar] [CrossRef] [PubMed]
- Oka, K.; Akamatsu, H.; Kodama, M.; Nakajima, H.; Kawada, T.; Otani, H. Host-specific AB-toxin production by germinating spores of Alternaria brassicicola is induced by a host-derived oligosaccharide. Physiol. Mol. Plant Pathol. 2005, 66, 12–19. [Google Scholar] [CrossRef]
- Schweizer, G.; Münch, K.; Mannhaupt, G.; Schirawski, J.; Kahmann, R.; Dutheil, J.Y. Positively selected effector genes and their contribution to virulence in the smut fungus Sporisorium reilianum. Genome Biol. Evol. 2018, 10, 629–645. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weiberg, A.; Dellota, E.; Yamane, D.; Jin, H. Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 2017, 14, 421–428. [Google Scholar] [CrossRef] [PubMed]



| Method 1 | Advantages | Disadvantages |
|---|---|---|
| Restriction fragment-length polymorphism(RFLP) | Can detect allelic variants | Large DNA quantity needed, typically only 1–3 loci detected, usually radioactive labeling is used |
| Random amplified polymorphic DNA (RAPD) | Faster than RFLP, less DNA is needed, can detect 1–10 variant loci, suitable for detection of broad scale genetic structural differences | Cannot detect allelic variants (heterozygous alleles or homologous alleles normally give the same result), less reliable, polymerase chain reaction (PCR)-dependent assay |
| Simple sequence repeats; microsatellites (SSR) | More accurate than RAPD, suitable for discriminating different subpopulations | Microsatellite markers may not be evenly distributed in the genome, SSR are located in non-coding regions, false alleles or null alleles may be detected due to technical artifacts, blurry bands may occur |
| Amplified fragment-length polymorphism (AFLP) | Combines benefits of RAPD and RFLP | Difficult to develop locus-specific marker (fragment) proprietary technology to score heterozygous and homozygous |
| Analysis of mitochondrial DNA (mtDNA analysis) | Powerful tool for studying inheritance of mitochondrial genomes, for phylogenetic and population genetic analysis, for species identification and barcoding | In uniparental-mtDNA inheritances, no information about other parent: should be coupled with genomic-DNA analyses. In case of mtDNA recombination (bi-parental inheritance) many analysis not doable |
| Sequencing of internal transcribed spacer regions (ITS sequencing) | ITS1 and ITS2 regions are species-specific and have large copy numbers, ITS sequencing can be used in metagenomics studies (meta-barcode), can be coupled with NGS technique | Limited to discriminate intra- and intergeneric species |
| Analysis of protein abundance of all proteins (proteomics) | Many different techniques available, e.g., two-dimensional electrophoresis coupled to mass spectrometric protein identification, can analyze vast array of proteins at once, can do high throughput, high sensitivity possible, relative as well as absolute protein abundance quantification possible | Each technique has its own limitation, not all proteins can be identified by one single method. Results may be tissue- and environmental condition-dependent |
| Sequencing using next-generation sequencing techniques (NGS sequencing) | Identify millions of single nucleotide polymorphisms (SNPs) as well as insertions and deletions (INDELs) at once | PCR-born false variants, data analysis needs bioinformatic know-how and computing power |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borah, N.; Albarouki, E.; Schirawski, J. Comparative Methods for Molecular Determination of Host-Specificity Factors in Plant-Pathogenic Fungi. Int. J. Mol. Sci. 2018, 19, 863. https://doi.org/10.3390/ijms19030863
Borah N, Albarouki E, Schirawski J. Comparative Methods for Molecular Determination of Host-Specificity Factors in Plant-Pathogenic Fungi. International Journal of Molecular Sciences. 2018; 19(3):863. https://doi.org/10.3390/ijms19030863
Chicago/Turabian StyleBorah, Nilam, Emad Albarouki, and Jan Schirawski. 2018. "Comparative Methods for Molecular Determination of Host-Specificity Factors in Plant-Pathogenic Fungi" International Journal of Molecular Sciences 19, no. 3: 863. https://doi.org/10.3390/ijms19030863
APA StyleBorah, N., Albarouki, E., & Schirawski, J. (2018). Comparative Methods for Molecular Determination of Host-Specificity Factors in Plant-Pathogenic Fungi. International Journal of Molecular Sciences, 19(3), 863. https://doi.org/10.3390/ijms19030863