Neuroprotective Effects of Four Phenylethanoid Glycosides on H2O2-Induced Apoptosis on PC12 Cells via the Nrf2/ARE Pathway
Abstract
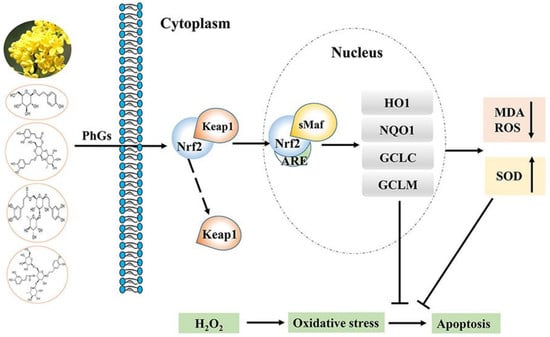
:1. Introduction
2. Results
2.1. PhGs Suppressed H2O2-Induced Cytotoxicity in PC12 Cells
2.2. PhGs Suppressed H2O2-Induced Intracellular Accumulation of ROS, Lipid Peroxidation (MDA), and Increased Superoxide Dismutase (SOD) Activities in PC12 Cells
2.3. PhGs Reversed H2O2-Induced Apoptosis in PC12 Cells
2.4. PhGs Alleviated H2O2-Induced Dysregulation of the Nrf2-ARE Pathways in PC12 Cells
2.5. PhGs Reversed H2O2-Induced Downregulation of Protein Expression of HO-1, NQO1, GCLC, and GCLM
2.6. Keap1 Expression and Molecular Docking Analysis
3. Discussion
4. Materials and Methods
4.1. Chemical Compounds and Reagents
4.2. Cell Culture
4.3. Cell Viability Assay
4.4. Apoptosis Assay
4.5. Measurement of Intracellular ROS, MDA Production, and SOD
4.6. Nrf2 Nuclear Translocation Immunofluorescence
4.7. Protein Extraction
4.8. Western Blot Analysis
4.9. Quantitative Real-Time PCR
4.10. siRNA Transfection
4.11. Molecular Docking Analysis
4.12. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GCLC | glutamate cysteine ligase-catalytic subunit |
| GCLM | glutamate cysteine ligase-catalytic modifier subunit |
| H2O2 | hydrogen peroxide |
| HO-1 | heme oxygenase 1 |
| Keap1 | Kelch ECH association protein 1 |
| NQO1 | NAD(P)H quinone oxidoreductase 1 |
| Nrf2 | nuclear factor erythroid 2-related factor 2 |
| PhGs | salidroside, acteoside, isoacteoside, and echinacoside |
| ROS | reactive oxygen species |
| ZnPP | zinc protoporphyrin |
References
- Sun, A.Y.; Chen, Y.M. Oxidative stress and neurodegenerative disorders. J. Biomed. Sci. 1998, 5, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, A.; Misro, M.M.; Aggarwal, A.; Sharma, R.K.; Nandan, D. Pathways involved in testicular germ cell apoptosis induced by H2O2 in vitro. FEBS J. 2009, 276, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Reactive oxygen species and the central nervous system. J. Neurochem. 1992, 59, 1609–1623. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.S.; Subbarao, K.V.; Ang, L.C. On the possible role of iron-induced free radical peroxidation in neural degeneration in Alzheimer’s disease. Ann. N. Y. Acad. Sci. 1992, 648, 326–327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab. Rev. 2006, 38, 769–789. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Tong, K.I.; Yamamoto, M. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic. Biol. Med. 2004, 36, 1208–1213. [Google Scholar] [CrossRef] [PubMed]
- Kong, A.N.; Owuor, E.; Yu, R.; Hebbar, V.; Chen, C.; Hu, R.; Mandlekar, S. Induction of xenobiotic enzymes by the map kinase pathway and the antioxidant or electrophile response element (ARE/EpRE). Drug Metab. Rev. 2001, 33, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Buendia, I.; Michalska, P.; Navarro, E.; Gameiro, I.; Egea, J.; León, R. Nrf2-ARE pathway: An emerging target against oxidative stress and neuroinflammation in neurodegenerative diseases. Pharmacol. Ther. 2016, 157, 84–104. [Google Scholar] [CrossRef] [PubMed]
- Skibinski, G.; Hwang, V.; Ando, D.M.; Daub, A.; Lee, A.K.; Ravisankar, A.; Modan, S.; Finucane, M.M.; Shaby, B.A.; Finkbeiner, S. Nrf2 mitigates LRRK2- and α-synuclein-induced neurodegeneration by modulating proteostasis. Proc. Natl. Acad. Sci. USA 2016, 114, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, C.; Riguera, R. Phenylethanoid glycosides in plants: Structure and biological activity. Nat. Prod. Rep. 1994, 11, 591–606. [Google Scholar] [CrossRef] [PubMed]
- Georgiev, M.; Alipieva, K.; Orhan, I.; Abrashev, R.; Denev, P.; Angelova, M. Antioxidant and cholinesterases inhibitory activities of Verbascum xanthophoeniceum Griseb. and its phenylethanoid glycosides. Food Chem. 2011, 128, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, J.; Ma, J.; Gu, Z.; Jiang, C.; Yu, L.; Fu, X. Neuroprotective Effects of Cistanches Herba Therapy on Patients with Moderate Alzheimer’s Disease. Evid.-Based Complement. Altern. Med. 2015, 2015, 103985. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; He, W.J.; Mo, L.; Shi, M.F.; Zhu, Y.Y.; Pan, S.; Li, X.R.; Xu, Q.M.; Yang, S.L. Antimicrobial, anti-inflammatory activities and toxicology of phenylethanoid glycosides from Monochasma savatieri Franch. ex Maxim. J. Ethnopharmacol. 2013, 149, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Yao, J.; Fang, J.N.; Ding, K. Structural characterization and immunological activity of two cold-water extractable polysaccharides from Cistanche deserticola Y. C. Ma. Carbohydr. Res. 2007, 342, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Mao, S.; Lu, B.; Yang, J.; Zhou, F.; Hu, Y.; Jiang, Y.; Shen, C.; Zhao, Y. Osmanthus fragrans Flower Extract and Acteoside Protect Against d-Galactose-Induced Aging in an ICR Mouse Model. J. Med. Food 2016, 19, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Mao, S.; Huang, W.; Lu, B.; Cai, Z.; Zhou, F.; Li, M.; Lou, T.; Zhao, Y. Phenylethanoid Glycoside Profiles and Antioxidant Activities of Osmanthus fragrans Lour. Flowers by UPLC/PDA/MS and Simulated Digestion Model. J. Agric. Food Chem. 2016, 64, 2459–2466. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ye, X.; Li, X.; Sun, X.; Liang, Q.; Tao, L.; Kang, X.; Chen, J. Salidroside protects against MPP+-induced apoptosis in PC12 cells by inhibiting the NO pathway. Brain Res. 2011, 1382, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ding, W.; Sun, H.; Zhou, Q.; Huang, J.; Li, X.; Xie, Y.; Chen, J. Salidroside protects PC12 cells from MPP⁺-induced apoptosis via activation of the PI3K/Akt pathway. Food Chem. Toxicol. 2012, 50, 2591–2597. [Google Scholar] [CrossRef] [PubMed]
- Sheng, G.Q.; Zhang, J.R.; Pu, X.P.; Ma, J.; Li, C.L. Protective effect of verbascoside on 1-methyl-4-phenylpyridinium ion-induced neurotoxicity in PC12 cells. Eur. J. Pharmacol. 2002, 451, 119–124. [Google Scholar] [CrossRef]
- Wang, H.; Xu, Y.; Yan, J.; Zhao, X.; Sun, X.; Zhang, Y.; Guo, J.; Zhu, C. Acteoside protects human neuroblastoma SH-SY5Y cells against beta-amyloid-induced cell injury. Brain Res. 2009, 1283, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Min, D.; Jin, Y.Z.; Yong, J.; Zheng, B.L.; Yao, H.W. Echinacoside rescues the SHSY5Y neuronal cells from TNFα-induced apoptosis. Chin. Pharmacol. Bull. 2005, 505, 11–18. [Google Scholar]
- Zhao, Q.; Gao, J.P.; Li, W.W.; Cai, D.F. Neurotrophic and neurorescue effects of Echinacoside in the subacute MPTP mouse model of Parkinson’s disease. Brain Res. 2010, 1346, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Koo, K.A.; Sung, S.H.; Park, J.H.; Kim, S.H.; Lee, K.Y.; Kim, Y.C. In vitro neuroprotective activities of phenylethanoid glycosides from Callicarpa dichotoma. Planta Med. 2005, 71, 778–780. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.G.; Li, X.; Xiong, C.; Yu, B.; Pu, X.; Ye, X.S. Synthetic phenylethanoid glycoside derivatives as potent neuroprotective agents. Eur. J. Med. Chem. 2015, 95, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Pang, H.; Wong, Y.H. Naturally occurring phenylethanoid glycosides: Potential leads for new therapeutics. Curr. Med. Chem. 2008, 15, 2592–2613. [Google Scholar] [CrossRef] [PubMed]
- Greene, L.A.; Tischler, A.S. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. USA 1976, 73, 2424–2428. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.N.; Sheng, Y.C.; Jiang, P.; Ji, L.L.; Xia, Y.Y.; Min, Y.; Wang, Z.T. The difference of glutathione antioxidant system in newly weaned and young mice liver and its involvement in isoline-induced hepatotoxicity. Arch. Toxicol. 2011, 85, 1267–1279. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Kang, M.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative Stress Sensor Keap1 Functions as an Adaptor for Cul3-Based E3 Ligase To Regulate Proteasomal Degradation of Nrf2. Mol. Cell. Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef] [PubMed]
- Cornelius, C.; Crupi, R.; Calabrese, V.; Graziano, A.; Milone, P.; Pennisi, G.; Radak, Z.; Calabrese, E.J.; Cuzzocrea, S. Traumatic brain injury: Oxidative stress and neuroprotection. Antioxid. Redox Signal. 2013, 19, 836–853. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Luo, W.; Sun, X.; Hao, S.; Zhang, Y.; Xu, F.; Wang, Z.; Liu, B. Hydrogen-rich saline protects against oxidative damage and cognitive deficits after mild traumatic brain injury. Brain Res. Bull. 2012, 88, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Ansari, M.A.; Roberts, K.N.; Scheff, S.W. A time course of contusion-induced oxidative stress and synaptic proteins in cortex in a rat model of TBI. J. Neurotrauma 2008, 25, 513–526. [Google Scholar] [CrossRef] [PubMed]
- Kuang, R.; Sun, Y.; Yuan, W.; Lei, L.; Zheng, X. Protective effects of echinacoside, one of the phenylethanoid glycosides, on H2O2-induced cytotoxicity in PC12 cells. Planta Med. 2009, 75, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Scapagnini, G.; Vasto, S.; Abraham, N.G.; Caruso, C.; Zella, D.; Fabio, G. Modulation of Nrf2/ARE pathway by food polyphenols: A nutritional neuroprotective strategy for cognitive and neurodegenerative disorders. Mol. Neurobiol. 2011, 44, 192–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerr, F.; Sofolaadesakin, O.; Ivanov, D.K.; Gatliff, J.; Gomez, P.B.; Bertrand, H.C.; Martinez, P.; Callard, R.; Snoeren, I.; Cochemé, H.M. Direct Keap1-Nrf2 disruption as a potential therapeutic target for Alzheimer’s disease. PLoS Genet. 2017, 13, e1006593. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.B.; Nielsen, S.E.; Berg, K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J. Immunol. Methods 1989, 119, 203–210. [Google Scholar] [CrossRef]
- Press, C. Virtual Screening in Drug Discovery; Crc Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Muegge, I.; Martin, Y.C.; Hajduk, P.J.; Fesik, S.W. Evaluation of PMF scoring in docking weak ligands to the FK506 binding protein. J. Med. Chem. 1999, 42, 2498–2503. [Google Scholar] [CrossRef] [PubMed]
- Kuntz, I.D.; Blaney, J.M.; Oatley, S.J.; Langridge, R.; Ferrin, T.E. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982, 161, 269–288. [Google Scholar] [CrossRef]
- MD, E.; Murray, C.W.; Auton, T.R.; Paolini, G.V.; Mee, R.P. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J. Comput.-Aided Mol. Des. 1997, 11, 425–445. [Google Scholar]









| Compounds Name | Total-Score | Crash | Polar | D-Score | PMF-Score | G-Score | Chem-Score | C-Score |
|---|---|---|---|---|---|---|---|---|
| Salidroside | 5.0951 | −1.0622 | 5.1831 | −561.448 | −30.2413 | −115.923 | −22.6459 | 4 |
| Acteoside | 7.4971 | −2.3674 | 7.8899 | −1562.04 | −67.978 | -192.18 | −33.947 | 5 |
| Isoacteoside | 8.1776 | −1.4 | 6.4762 | −1439.24 | −88.9989 | −155.055 | −26.1735 | 4 |
| Echinacoside | 9.3402 | −2.6794 | 9.6386 | −1756.32 | −82.9345 | −187.808 | −32.4387 | 4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Xu, T.; Zhou, F.; Wang, M.; Song, H.; Xiao, X.; Lu, B. Neuroprotective Effects of Four Phenylethanoid Glycosides on H2O2-Induced Apoptosis on PC12 Cells via the Nrf2/ARE Pathway. Int. J. Mol. Sci. 2018, 19, 1135. https://doi.org/10.3390/ijms19041135
Li M, Xu T, Zhou F, Wang M, Song H, Xiao X, Lu B. Neuroprotective Effects of Four Phenylethanoid Glycosides on H2O2-Induced Apoptosis on PC12 Cells via the Nrf2/ARE Pathway. International Journal of Molecular Sciences. 2018; 19(4):1135. https://doi.org/10.3390/ijms19041135
Chicago/Turabian StyleLi, Maiquan, Tao Xu, Fei Zhou, Mengmeng Wang, Huaxin Song, Xing Xiao, and Baiyi Lu. 2018. "Neuroprotective Effects of Four Phenylethanoid Glycosides on H2O2-Induced Apoptosis on PC12 Cells via the Nrf2/ARE Pathway" International Journal of Molecular Sciences 19, no. 4: 1135. https://doi.org/10.3390/ijms19041135
APA StyleLi, M., Xu, T., Zhou, F., Wang, M., Song, H., Xiao, X., & Lu, B. (2018). Neuroprotective Effects of Four Phenylethanoid Glycosides on H2O2-Induced Apoptosis on PC12 Cells via the Nrf2/ARE Pathway. International Journal of Molecular Sciences, 19(4), 1135. https://doi.org/10.3390/ijms19041135