A Review of the Interactions between Wheat and Wheat Pathogens: Zymoseptoria tritici, Fusarium spp. and Parastagonospora nodorum
Abstract
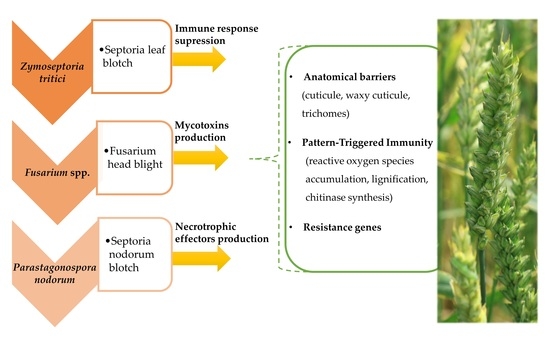
:1. Introduction
2. The Infection Cycle of the Hemibiotrophic Fungi Zymoseptoria tritici and Fusarium spp. and the Necrotrophic Fungus Parastagonospora nodorum
3. The Role of Morphological and Anatomical Barriers in Conditioning Resistance to Pathogens in Wheat
4. Pathogen-Induced Resistance in Plants–Pattern-Triggered Immunity
5. Gene Expression in Infected Wheat
6. Characteristics of Loci in the Wheat Genome Conferring Resistance to Pathogenic Infections
7. Practical Application Wheat Defense Mechanisms
8. Conclusions
Author Contributions
Conflicts of Interest
Abbreviations
| AM | Association mapping |
| AOX | Alternative oxidase |
| Cas | CRISPR associated |
| CEBIP | Chitin elicitor-binding protein |
| CERK1 | Chitin elicitor response kinase 1 |
| CRISPR | Clustered Regularly-Interspaced Short Palindromic Repeats |
| DArT | Diversity Arrays Technology |
| DMI | Demethylation inhibitors |
| DON | Deoxynivalenol |
| ETI | Effector-triggered immunity |
| FHB | Fusarium head blight |
| HIGS | Host-induced gene silencing |
| HR | Hypersensitive response |
| LOX | Lipoxygenase |
| LysM | Lysine motif |
| MAPK | Mitogen-activated protein kinase |
| MeJA | Methyl-jasmonate |
| NEP1 | Necrosis and ethylene-inducing peptide |
| NIP | Necrosis-inducing proteins |
| NRPSs | Nonribosomal peptide synthetases |
| PAL | Phenylalanine ammonia-lyase |
| PCD | Programed cell death |
| PKSs | Polyketide synthases |
| PR | Pathogenesis-related |
| PTI | Pattern-triggered immunity |
| QoI | Quinone outside inhibitors |
| QTL | Quantitative trait loci |
| RILs | Recombinant inbred lines |
| ROS | Reactive oxygen species |
| SA | Salicylic acid |
| SIGS | Spray-induced gene silencing |
| STB | Septoria leaf blotch |
| TCA | Tricarboxylic acid |
| TGS | Transcriptional gene silencing |
| UGT | UDP-glucuronosyltransferase |
| VIGS | Virus-induced gene silencing |
References
- Fedak, G. Alien Introgressions from wild Triticum species, T. monococcum, T. urartu, T. turgidum, T. dicoccum, T. dicoccoides, T. carthlicum, T. araraticum, T. timopheevii, and T. miguschovae. In Alien Introgression in Wheat; Molnar, M., Ceoloni, C., Doležel, J., Eds.; Springer International Publishing: Basel, Switzerland, 2015; pp. 191–219. ISBN 978-3-319-23493-9. [Google Scholar]
- Food and Agriculture Organization of the United Nations (FAO). Record-Breaking Production to Lift World Cereal Stocks to An-All Time High in 2017/18, Despite Growing Consumption and Robust Trade. Available online: http://www.fao.org/worldfoodsituation/csdb/en/ (accessed on 3 November 2017).
- Główny Urząd Statystyczny (GUS). Rolnictwo w 2016 r; Główny Urząd Statystyczny Departament Rolnictwa: Warszawa, Poland, 2017; pp. 45–62. ISBN 1507-9724.
- Çifçi, E.A.; Yağdi, K. Study of genetic diversity in wheat (Triticum aestivum) varieties using random amplified polymorphic DNA (RAPD) analysis. Turk. J. Field Crops 2012, 17, 91–95. [Google Scholar]
- Rudd, J.J. Previous bottlenecks and future solutions to dissecting the Zymoseptoria tritici–wheat host-pathogen interaction. Fungal Genet. Biol. 2015, 79, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Heick, T.M.; Justesen, A.F.; Jørgensen, L.N. Anti-resistance strategies for fungicides against wheat pathogen Zymoseptoria tritici with focus on DMI fungicides. Crop Prot. 2017, 99, 108–117. [Google Scholar] [CrossRef]
- Stuper-Szablewska, K.; Kurasiak-Popowska, D.; Nawracala, J.; Perkowski, J. Wheat grain contaminated by microscopic fungi as a source of phenolic acids for potential application in the pharmaceutical industry. Przem. Chem. 2016, 95, 1233–1236. [Google Scholar]
- Simón, M.R.; Perelló, A.E.; Cordo, C.A.; Struik, P.C. Influence of Septoria tritici on yield, yield components, and test weight of wheat under two nitrogen fertilization conditions. Crop Sci. 2002, 42, 1974–1981. [Google Scholar] [CrossRef]
- Berg, F.; Paveley, N.D.; Bosch, F. Dose and number of applications that maximize fungicide effective life exemplified by Zymoseptoria tritici on wheat—A model analysis. Plant Pathol. 2016, 65, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Wegulo, S.N.; Bockus, W.W.; Nopsa, J.H.; De Wolf, E.D.; Eskridge, K.M.; Peiris, K.H.; Dowell, F.E. Effects of integrating cultivar resistance and fungicide application on Fusarium head blight and deoxynivalenol in winter wheat. Plant Dis. 2011, 95, 554–560. [Google Scholar] [CrossRef]
- Wegulo, S.N.; Zwingman, M.V.; Breathnach, J.A.; Baenziger, P.S. Economic returns from fungicide application to control foliar fungal diseases in winter wheat. Crop Prot. 2011, 30, 685–692. [Google Scholar] [CrossRef]
- Franke, J.; Menz, G. Multi-temporal wheat disease detection by multi-spectral remote sensing. Precis. Agric. 2007, 8, 161–172. [Google Scholar] [CrossRef]
- Lenc, L.; Wyczling, D.; Sadowski, C. Zasiedlenie ziarna pszenicy ozimej przez grzyby rodzaju Fusarium w zależności od przedplonu, uprawianej odmiany i stosowanych fungicydów. Ochrona Ś Rodowiska i Zasobów Naturalnych 2009, 41, 563–571. [Google Scholar]
- D’Angelo, D.L.; Bradley, C.A.; Ames, K.A.; Willyerd, K.T.; Madden, L.V.; Paul, P.A. Efficacy of fungicide applications during and after anthesis against Fusarium head blight and deoxynivalenol in soft red winter wheat. Plant Dis. 2014, 98, 1387–1397. [Google Scholar] [CrossRef]
- COBORU. Lista Odmian Zalecanych (Loz) Na 2016 Rok Pszenica Ozima. Available online: http://www.coboru.pl/PlikiWynikow/19_2016_LZO_7_ZYZO.pdf (accessed on 3 January 2018).
- Yang, F.; Li, W.; Derbyshire, M.; Larsen, M.R.; Rudd, J.J.; Palmisano, G. Unraveling incompatibility between wheat and the fungal pathogen Zymoseptoria tritici through apoplastic proteomics. BMC Genom. 2015, 16, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, L.; Eyal, Z. The histology of processes associated with the infection of resistant and susceptible wheat cultivars with Septoria tritici. Plant Pathol. 1993, 42, 737–743. [Google Scholar] [CrossRef]
- Yang, F.; Li, W.; Jørgensen, H.J.L. Transcriptional Reprogramming of Wheat and the Hemibiotrophic Pathogen Septoria tritici during Two Phases of the Compatible Interaction. PLoS ONE 2013, 8, e81606. [Google Scholar] [CrossRef] [PubMed]
- Kettles, G.J.; Kanyuka, K. Dissecting the Molecular Interactions between Wheat and the Fungal Pathogen Zymoseptoria tritici. Front. Plant Sci. 2016, 7, 508. [Google Scholar] [CrossRef] [PubMed]
- Rohel, E.A.; Payne, A.C.; Fraaije, B.A.; Hollomon, D.W. Exploring infection of wheat and carbohydrate metabolism in Mycosphaerella graminicola transformants with differentially regulated green fluorescent protein expression. Mol. Plant Microbe Interact. 2001, 14, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Palma-Guerrero, J.; Torriani, S.F.; Zala, M.; Carter, D.; Courbot, M.; Rudd, J.J.; McDonald, B.A.; Croll, D. Comparative transcriptomic analyses of Zymoseptoria tritici strains show complex lifestyle transitions and intraspecific variability in transcription profiles. Mol. Plant Pathol. 2016, 17, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.; Kombrink, A.; Motteram, J.; Loza-Reyes, E.; Lucas, J.; Hammond-Kosack, K.E.; Thomma, B.P.H.J.; Rudd, J.J. Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 2011, 156, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, G. Cell biology of Zymoseptoria tritici: Pathogen cell organization and wheat infection. Fungal Genet. Biol. 2015, 79, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Rudd, J.J.; Hammond-Kosack, K.E.; Kanyuka, K. Mycosphaerella graminicola LysM effector-mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat. Mol. Plant Microbe Interact. 2014, 27, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Sella, L.; Gazzetti, K.; Faoro, F.; Odorizzi, S.; D’Ovidio, R.; Schäfer, W.; Favaron, F. A Fusarium graminearum xylanase expressed during wheat infection is a necrotizing factor but is not essential for virulence. Plant Physiol. Biochem. 2013, 64, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Boenisch, M.J.; Schäfer, W. Fusarium graminearum forms mycotoxin producing infection structures on wheat. BMC Plant Biol. 2011, 11, 110. [Google Scholar] [CrossRef] [PubMed]
- Dyer, R.B.; Plattner, R.D.; Kendra, D.F.; Brown, D.W. Fusarium graminearum TRI14 is required for high virulence and DON production on wheat but not for DON synthesis in vitro. J. Agric. Food Chem. 2005, 53, 9281–9287. [Google Scholar] [CrossRef] [PubMed]
- Stephens, A.E.; Gardiner, D.M.; White, R.G.; Munn, A.L.; Manners, J.M. Phases of infection and gene expression of Fusarium graminearum during crown rot disease of wheat. Mol. Plant Microbe Interact. 2008, 21, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Jenczmionka, N.J.; Maier, F.J.; Lösch, A.P.; Schäfer, W. Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head-blight disease of wheat, are regulated by the MAP kinase gpmk1. Curr. Genet. 2003, 43, 87–95. [Google Scholar] [PubMed]
- Lu, S.W.; Kroken, S.; Lee, B.N.; Robbertse, B.; Churchill, A.C.; Yoder, O.C.; Turgeon, B.G. A novel class of gene controlling virulence in plant pathogenic ascomycete fungi. Proc. Natl. Acad. Sci. USA 2003, 100, 5980–5985. [Google Scholar] [CrossRef] [PubMed]
- Bluhm, B.H.; Zhao, X.; Flaherty, J.E.; Xu, J.R.; Dunkle, L.D. RAS2 regulates growth and pathogenesis in Fusarium graminearum. Mol. Plant Microbe Interact. 2007, 20, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Voigt, C.A.; Schäfer, W.; Salomon, S. A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J. 2005, 42, 364–375. [Google Scholar] [CrossRef] [PubMed]
- IpCho, S.V.; Tan, K.C.; Koh, G.; Gummer, J.; Oliver, R.P.; Trengove, R.D.; Solomon, P.S. The transcription factor StuA regulates central carbon metabolism, mycotoxin production, and effector gene expression in the wheat pathogen Stagonospora nodorum. Eukaryot. Cell 2010, 9, 1100–1108. [Google Scholar] [CrossRef] [PubMed]
- Rybak, K.; See, P.T.; Phan, H.T.T.; Syme, R.A.; Moffat, C.S.; Oliver, R.P.; Tan, K.C. A functionally conserved Zn2Cys6 binuclear cluster transcription factor class regulates necrotrophic effector gene expression and host-specific virulence of two major Pleosporales fungal pathogens of wheat. Mol. Plant Pathol. 2017, 18, 420–434. [Google Scholar] [CrossRef] [PubMed]
- Faris, J.D.; Anderson, J.A.; Francl, L.J.; Jordahl, J.G. Chromosomal location of a gene conditioning insensitivity in wheat to a necrosis-inducing culture filtrate from Pyrenophora tritici-repentis. Phytopathology 1996, 86, 459–463. [Google Scholar] [CrossRef]
- Ipcho, S.V.; Hane, J.K.; Antoni, E.A.; Ahren, D.A.G.; Henrissat, B.; Friesen, T.L.; Solomon, P.S.; Oliver, R.P. Transcriptome analysis of Stagonospora nodorum: Gene models, effectors, metabolism and pantothenate dispensability. Mol. Plant Pathol. 2012, 13, 531–545. [Google Scholar] [CrossRef] [PubMed]
- Hane, J.K.; Lowe, R.G.; Solomon, P.S.; Tan, K.C.; Schoch, C.L.; Spatafora, J.W.; Crous, P.W.; Kodira, C.; Birren, B.W.; Galagan, J.E.; et al. Dothideomycete—Plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum. Plant Cell 2007, 19, 3347–3368. [Google Scholar] [CrossRef] [PubMed]
- Lehtinen, U. Plant cell wall degrading enzymes of Septoria nodorum. Physiol. Mol. Plant Pathol. 1993, 43, 121–134. [Google Scholar] [CrossRef]
- Cairns, T.; Meyer, V. In silico prediction and characterization of secondary metabolite biosynthetic gene clusters in the wheat pathogen Zymoseptoria tritici. BMC Genom. 2017, 18, 631. [Google Scholar] [CrossRef] [PubMed]
- Palma-Guerrero, J.; Ma, X.; Torriani, S.F.; Zala, M.; Francisco, C.S.; Hartmann, F.E.; Croll, D.; McDonald, B.A. Comparative transcriptome analyses in Zymoseptoria tritici reveal significant differences in gene expression among strains during plant infection Mol. Plant Microbe Interact. 2017, 30, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewska, H.; Surma, M.; Krystkowiak, K.; Adamski, T.; Kuczyńska, A.; Ogrodowicz, P.; Mikołajczak, K.; Belter, J.; Majka, M.; Kaczmarek, Z.; et al. Simultaneous selection for yield-related traits and susceptibility to Fusarium head blight in spring wheat RIL population. Breed. Sci. 2016, 66, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Wu, B.; Li, Q.; Du, X.; Jia, K. Monitoring crop phenology with MERIS data—A case study of winter wheat in North China Plain. In Proceedings of the Progress in Electromagnetics Research Symposium, Beijing, China, 23–27 March 2009; pp. 1225–1228. [Google Scholar]
- Tschanz, A.T.; Horst, R.K.; Nelson, P.E. The effect of environment on sexual reproduction of Gibberella zeae. Mycologia 1976, 68, 327–340. [Google Scholar] [CrossRef]
- Schmale, D.G., III; Bergstrom, G.C. The aerobiology and population genetic structure of Gibberella zeae. Phytopathology 2005, 95. [Google Scholar] [CrossRef]
- Manstretta, V.; Rossi, V. Effects of temperature and moisture on development of Fusarium graminearum perithecia in maize stalk residues. Appl. Environ. Microbiol. 2016, 82, 184–191. [Google Scholar] [CrossRef] [PubMed]
- David, R.F.; Marr, L.C.; Schmale, D.G. Ascospore release and discharge distances of Fusarium graminearum under controlled temperature and relative humidity. Eur. J. Plant Pathol. 2016, 146, 59–69. [Google Scholar] [CrossRef]
- Saccon, F.A.; Elrewainy, A.; Parcey, D.; Paliwal, J.; Sherif, S.S. Detection of Fusarium on Wheat using near infrared hyperspectral imaging. In Proceedings of the Photonics North (PN), Quebec City, QC, Canada, 24–26 May 2016. [Google Scholar]
- Xu, X.; Nicholson, P. Community ecology of fungal pathogens causing wheat head blight. Annu. Rev. Phytopathol. 2009, 47, 83–103. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Buchenauer, H. Ultrastructural and immunocytochemical investigation of pathogen development and host responses in resistant and susceptible wheat spikes infected by Fusarium culmorum. Physiol. Mol. Plant Pathol. 2000, 57, 255–268. [Google Scholar] [CrossRef]
- Chetouhi, C.; Bonhomme, L.; Lasserre-Zuber, P.; Cambon, F.; Pelletier, S.; Renou, J.P.; Langin, T. Transcriptome dynamics of a susceptible wheat upon Fusarium head blight reveals that molecular responses to Fusarium graminearum infection fit over the grain development processes. Funct. Integr. Genom. 2016, 16, 183–201. [Google Scholar] [CrossRef] [PubMed]
- Zinkernagel, V.; Reiss, F.; Wendland, M. Infection structures of Septoria nodorum in leaves of susceptible wheat cultivars. J. Plant Dis. Prot. 1988, 95, 169–175. [Google Scholar]
- Solomon, P.S.; Lowe, R.G.T.; Tan, K.C.; Waters, O.D.C.; Oliver, R.P. Stagonospora nodorum: Cause of Stagonospora nodorum blotch of wheat. Mol. Plant Pathol. 2006, 7, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Sommerhalder, R.J.; McDonald, B.A.; Mascher, F.; Zhan, J. Sexual recombinants make a significant contribution to epidemics caused by the wheat pathogen Phaeosphaeria nodorum. Phytopathology 2010, 100, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Estep, L.K.; Torriani, S.F.F.; Zala, M.; Anderson, N.P.; Flowers, M.D.; McDonald, B.A.; Mundt, C.C.; Brunner, P.C. Emergence and early evolution of fungicide resistance in North American populations of Zymoseptoria tritici. Plant Pathol. 2015, 64, 961–971. [Google Scholar] [CrossRef]
- Pöggeler, S.; Wöstemeyer, J. The Mycota, Volume 14: Evolution of Fungi and Fungal-Like Organisms; Pöggeler, S., Wöstemeyer, J., Eds.; Springer: Berlin, Germany, 2011; pp. 223–226, ISBN-13 9783642199738. [Google Scholar]
- McDonald, M.C.; Ahren, D.; Simpfendorfer, S.; Milgate, A.; Solomon, P.S. The discovery of the virulence gene ToxA in the wheat and barley pathogen Bipolaris sorokiniana. Mol. Plant Pathol. 2018, 19, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Winterberg, B.; Du Fall, L.A.; Song, X.; Pascovici, D.; Care, N.; Molloy, M.; Ohms, S.; Solomon, P.S. The necrotrophic effector protein SnTox3 re-programs metabolism and elicits a strong defence response in susceptible wheat leaves. BMC Plant Biol. 2014, 14, 215. [Google Scholar] [CrossRef] [PubMed]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.; Barthlott, W.; Koch, S.; Hommes, A.; Wandelt, K.; Mamdouh, W.; De-Feyter, S.; Broekmann, P. Structural analysis of wheat wax (Triticum aestivum, c.v. ‘Naturastar’ L.): From the molecular level to three dimensional crystals. Planta 2006, 223, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, J.R.; Fish, L.J.; Leverington-Waite, M.A.; Wang, Y.; Howell, P.; Snape, J.W. Mapping of a gene (Vir) for a non-glaucous, viridescent phenotype in bread wheat derived from Triticum dicoccoides, and its association with yield variation. Euphytica 2008, 159, 333–341. [Google Scholar] [CrossRef]
- Wicki, W.; Winzeler, M.; Schmid, J.E.; Stamp, P.; Messmer, M. Inheritance of resistance to leaf and glume blotch caused by Septoria nodorum Berk. in winter wheat. Theor. Appl. Genet. 1999, 99, 1265–1272. [Google Scholar] [CrossRef]
- Gunnaiah, R.; Kushalappa, A.C.; Duggavathi, R.; Fox, S.; Somers, D.J. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against F. graminearum. PLoS ONE 2012, 7, e40695. [Google Scholar] [CrossRef] [PubMed]
- Freeman, B.C.; Beattie, G.A. An overview of plant defenses against pathogens and herbivores. Plant Health Instr. 2008, 94. [Google Scholar] [CrossRef]
- Fones, H.N.; Eyles, C.J.; Kay, W.; Cowper, J.; Gurr, S.J. A role for random, humidity-dependent epiphytic growth prior to invasion of wheat by Zymoseptoria tritici. Fungal Genet. Biol. 2017, 106, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Zelinger, E.; Hawes, C.R.; Gurr, S.J.; Dewey, F.M. Attachment and adhesion of conidia of Stagonospora nodorum to natural and artificial surfaces. Physiol. Mol. Plant Pathol. 2006, 68, 209–215. [Google Scholar] [CrossRef]
- Duncan, K.E.; Howard, R.J. Cytological analysis of wheat infection by the leaf blotch pathogen Mycosphaerella graminicola. Mycol. Res. 2000, 104, 1074–1082. [Google Scholar] [CrossRef]
- Shetty, N.P.; Kristensen, B.K.; Newman, M.A.; Møller, K.; Gregersen, P.L.; Jørgensen, H.L. Association of hydrogen peroxide with restriction of Septoria tritici in resistant wheat. Physiol. Mol. Plant Pathol. 2003, 62, 333–346. [Google Scholar] [CrossRef]
- Kema, G.H.J.; Sayoud, R.; Annone, J.G.; Van Silfhout, C.H. Genetic variation for virulence and resistance in the wheat-Mycosphaerella graminicola pathosystem. II: Analysis of interactions between pathogen isolates and host cultivars. Phytopathology 1996, 86, 213–220. [Google Scholar] [CrossRef]
- Pritsch, C.; Muehlbauer, G.J.; Bushnell, W.R.; Somers, D.A.; Vance, C.P. Fungal development and induction of defense response genes during early infection of wheat spikes by Fusarium graminearum. Mol. Plant Microbe Interact. 2000, 13, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Kaku, H.; Shibuya, N. Molecular mechanisms of chitin recognition and immune signaling by LysM-receptors. Physiol. Mol. Plant Pathol. 2016, 95, 60–65. [Google Scholar] [CrossRef]
- Altenbach, D.; Robatzek, S. Pattern recognition receptors: From the cell surface to intracellular dynamics. Mol. Plant Microbe Interact. 2007, 20, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Keller, B.; McDonald, B.A.; Palma-Guerrero, J.; Wicker, T. Comparative transcriptomics reveals how wheat responds to infection by Zymoseptoria tritici. Mol. Plant Microbe Interact. 2018, 31, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Shetty, N.P.; Jensen, J.D.; Knudsen, A.; Finnie, C.; Geshi, N.; Blennow, A.; Collinge, D.B.; Jørgensen, H.J.L. Effects of β-1,3-glucan from Septoria tritici on structural defence responses in wheat. J. Exp. Bot. 2009, 60, 4287–4300. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Du, Y.; Dong, Z. Chitin oligosaccharide and chitosan oligosaccharide: Two similar but different plant elicitors. Front. Plant Sci. 2016, 7, 522. [Google Scholar] [CrossRef] [PubMed]
- Moravčíková, J.; Ujvariová, N.; Žur, I.; Gálová, Z.; Gregorová, Z.; Zimová, M.; Boszorádová, E.; Matušíková, I. Chitinase Activities in Wheat and Its Relative Species. Agriculture (Polnohospodárstvo) 2017, 63, 14–22. [Google Scholar] [CrossRef]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Zhang, X.C.; Neece, D.; Ramonell, K.M.; Clough, S.; Kim, S.Y.; Stacey, M.G.; Stacey, G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 2008, 20, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Buchenauer, H.; Huang, L.; Han, Q.; Zhang, H. Cytological and immunocytochemical studies on responses of wheat spikes of the resistant Chinese cv. Sumai 3 and the susceptible cv. Xiaoyan 22 to infection by Fusarium graminearum. Eur. J. Plant Pathol. 2008, 120, 383–396. [Google Scholar] [CrossRef]
- Xiao, J.; Jin, X.; Jia, X.; Wang, H.; Cao, A.; Zhao, W.; Pei, H.; Xue, Z.; He, L.; Chen, Q.; et al. Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genom. 2013, 14, 197. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vallet, A.; Mesters, J.R.; Thomma, B.P. The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 2015, 39, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.; Farkaš, V.; Sanz, A.B.; Cabib, E. Strengthening the fungal cell wall through chitin–glucan cross-links: Effects on morphogenesis and cell integrity. Cell. Microbiol. 2016, 18, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Tan, Y.; Liu, N.; Liao, Y.; Sun, C.; Wang, S.; Wu, A. Functional Agents to Biologically Control Deoxynivalenol Contamination in Cereal Grains. Front. Microbiol. 2016, 7, 395. [Google Scholar] [CrossRef] [PubMed]
- Walter, S.; Kahla, A.; Arunachalam, C.; Perochon, A.; Khan, M.R.; Scofield, S.R.; Doohan, F.M. A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J. Exp. Bot. 2015, 66, 2583–2593. [Google Scholar] [CrossRef] [PubMed]
- Góral, T.; Ochodzki, P.; Walentyn-Góral, D.; Belter, J.; Majka, M.; Kwiatek, M.; Wiśniewska, H.; Bogacki, J.; Drzazga, T.; Ługowska, B.; et al. Odporność genotypów pszenicy ozimej na fuzariozę kłosów i akumulację toksyn fuzaryjnych w ziarnie scharakteryzowana za pomocą różnych typów odporności. [Resistance of winter wheat lines to Fusarium head blight and Fusarium toxins accumulation characterized using different types of resistance]. Biuletyn Instytutu Hodowli i Aklimatyzacji Roślin 2015, 276, 19–37. [Google Scholar]
- Lemmens, M.; Scholz, U.; Berthiller, F.; Dall’Asta, C.; Koutnik, A.; Schuhmacher, R.; Gerhard, A.; Buerstmayr, H.; Mesterházy, A.; Krska, R.; et al. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol. Plant Microbe Interact. 2005, 18, 1318–1324. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, D.M.; Kazan, K.; Praud, S.; Torney, F.J.; Rusu, A.; Manners, J.M. Early activation of wheat polyamine biosynthesis during Fusarium head blight implicates putrescine as an inducer of trichothecene mycotoxin production. BMC Plant Biol. 2010, 10, 289. [Google Scholar] [CrossRef] [PubMed]
- Rudd, J.J.; Kanyuka, K.; Hassani-Pak, K.; Derbyshire, M.; Andongabo, A.; Devonshire, J.; Lysenko, A.; Saqi, M.; Desai, N.M.; Powers, S.J.; et al. Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 2015, 167, 1158–1185. [Google Scholar] [PubMed]
- Shetty, N.P.; Mehrabi, R.; Lutken, H.; Haldrup, A.; Kema, G.H.; Collinge, D.B.; Jørgensen, H.J. Role of hydrogen peroxide during the interaction between the hemibiotrophic fungal pathogen Septoria tritici and wheat. New Phytol. 2007, 174, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Shetty, N.P.; Jørgensen, H.J.L.; Jensen, J.D.; Collinge, D.B.; Shetty, H.S. Roles of reactive oxygen species in interactions between plants and pathogens. Eur. J. Plant Pathol. 2008, 121, 267–280. [Google Scholar] [CrossRef]
- Ray, S.; Anderson, J.M.; Urmeev, F.I.; Goodwin, S.B. Rapid induction of a protein disulfide isomerase and defense-related genes in wheat in response to the hemibiotrophic fungal pathogen Mycosphaerella graminicola. Plant Mol. Biol. 2003, 53, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Keon, J.; Antoniw, J.; Carzaniga, R.; Deller, S.; Ward, J.L.; Baker, J.M.; Beale, M.H.; Hammond-Kosack, K.; Rudd, J.J. Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant Microbe Interact. 2007, 20, 178–193. [Google Scholar] [CrossRef] [PubMed]
- Rudd, J.J.; Keon, J.; Hammond-Kosack, K.E. The Wheat mitogen- activated protein kinases TaMPK3 and TaMPK6 are differentially regulated at multiple levels during compatible disease interactions with Mycosphaerella graminicola. Plant Physiol. 2008, 147, 802–815. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Sato, I.; Ishizaka, M.; Yoshida, S.I.; Koitabashi, M.; Yoshida, S.; Tsushima, S. Bacterial cytochrome P450 system catabolizing the Fusarium toxin deoxynivalenol. Appl. Environ. Microbiol. 2013, 79, 1619–1628. [Google Scholar] [CrossRef] [PubMed]
- Berthiller, F.; Dall’Asta, C.; Schuhmacher, R.; Lemmens, M.; Adam, G.; Krska, R. Masked mycotoxins: Determination of a deoxynivalenol glucoside in artificially and naturally contaminated wheat by liquid chromatography—Tandem mass spectrometry. J. Agric. Food Chem. 2005, 53, 3421–3425. [Google Scholar] [CrossRef] [PubMed]
- Berthiller, F.; Dall’Asta, C.; Corradini, R.; Marchelli, R.; Sulyok, M.; Krska, R.; Adam, G.; Schuhmacher, R. Occurrence of deoxynivalenol and its 3-β-d-glucoside in wheat and maize. Food Addit. Contam. 2009, 26, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Friesen, T.L.; Zhang, Z.; Solomon, P.S.; Oliver, R.P.; Faris, J.D. Characterization of the interaction of a novel Stagonospora nodorum host-selective toxin with a wheat susceptibility gene. Plant Physiol. 2008, 146, 682–693. [Google Scholar] [CrossRef] [PubMed]
- Faris, J.D.; Zhang, Z.; Lu, H.; Lu, S.; Reddy, L.; Cloutier, S.; Fellers, J.P.; Meinhardt, S.W.; Rasmussen, J.B.; Xu, S.S.; et al. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. USA 2010, 107, 13544–13549. [Google Scholar] [CrossRef] [PubMed]
- Oliver, R.P.; Friesen, T.L.; Faris, J.D.; Solomon, P.S. Stagonospora nodorum: From pathology to genomics and host resistance. Annu. Rev. Phytopathol. 2012, 50, 23–43. [Google Scholar] [CrossRef] [PubMed]
- Abeysekara, N.S.; Friesen, T.L.; Keller, B.; Faris, J.D. Identification and characterization of a novel host–toxin interaction in the wheat–Stagonospora nodorum pathosystem. Theor. Appl. Genet. 2009, 120, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.H.; Faris, J.D.; Meinhardt, S.W.; Ali, S.; Rasmussen, J.B.; Friesen, T.L. Genetic and physical mapping of a gene conditioning sensitivity in wheat to a partially purified host-selective toxin produced by Stagonospora nodorum. Phytopathology 2004, 94, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Dickman, M.B.; Park, Y.K.; Oltersdorf, T.; Li, W.; Clemente, T.; French, R. Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc. Natl. Acad. Sci. USA 2001, 98, 6957–6962. [Google Scholar] [CrossRef] [PubMed]
- Manning, V.A.; Ciuffetti, L.M. Localization of Ptr ToxA produced by Pyrenophora tritici-repentis reveals protein import into wheat mesophyll cells. Plant Cell 2005, 17, 3203–3212. [Google Scholar] [CrossRef] [PubMed]
- Friesen, T.L.; Meinhardt, S.W.; Faris, J.D. The Stagonospora nodorum-wheat pathosystem involves multiple proteinaceous host-selective toxins and corresponding host sensitivity genes that interact in an inverse gene-for-gene manner. Plant J. 2007, 51, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Friesen, T.L.; Xu, S.S.; Shi, G.; Liu, Z.; Rasmussen, J.B.; Faris, J.D. Two putatively homoeologous wheat genes mediate recognition of SnTox3 to confer effector-triggered susceptibility to Stagonospora nodorum. Plant J. 2011, 65, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.B.; Thompson, I. Development of isogenic lines for resistance to Septoria tritici blotch in wheat. Czech J. Genet. Plant 2011, 47, 98–101. [Google Scholar] [CrossRef]
- Brown, J.K.; Chartrain, L.; Lasserre-Zuber, P.; Saintenac, C. Genetics of resistance to Zymoseptoria tritici and applications to wheat breeding. Fungal Genet. Biol. 2015, 79, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Kollers, S.; Rodemann, B.; Ling, J.; Korzun, V.; Ebmeyer, E.; Argillier, O.; Hinze, M.; Plieske, J.; Kulosa, D.; Ganal, M.W.; et al. Genetic architecture of resistance to Septoria tritici blotch (Mycosphaerella graminicola) in European winter wheat. Mol. Breed. 2013, 32, 411–423. [Google Scholar] [CrossRef]
- Adhikari, T.B.; Jackson, E.W.; Gurung, S.; Hansen, J.M.; Bonman, J.M. Association mapping of quantitative resistance to Phaeosphaeria nodorum in spring wheat landraces from the USDA National Small Grains Collection. Phytopathology 2011, 101, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Ghaffary, S.M.T.; Robert, O.; Laurent, V.; Lonnet, P.; Margalé, E.; van der Lee, T.A.; Visser, R.G.F.; Kema, G.H. Genetic analysis of resistance to Septoria tritici blotch in the French winter wheat cultivars Balance and Apache. Theor. Appl. Genet. 2011, 123, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Raman, R.; Milgate, A.W.; Imtiaz, M.; Tan, K.; Raman, H. Molecular mapping and physical location of major gene conferring seedling resistance to Septoria tritici blotch in wheat. Mol. Breed. 2009, 24, 153–164. [Google Scholar] [CrossRef]
- McCartney, C.A.; Brule-Babel, A.L.; Lamari, L.; Somers, D.J. Chromosomal location of a race-specific resistance gene to Mycosphaerella graminicola in the spring wheat ST6. Theor. Appl. Genet. 2003, 107, 1181–1186. [Google Scholar] [CrossRef] [PubMed]
- Ghaffary, S.M.T.; Faris, J.D.; Friesen, T.L.; Visser, R.G.; van der Lee, T.A.; Robert, O.; Kema, G.H. New broad-spectrum resistance to Septoria tritici blotch derived from synthetic hexaploid wheat. Theor. Appl. Genet. 2012, 124, 125–142. [Google Scholar] [CrossRef] [PubMed]
- Cuthbert, P.A.; Somers, D.J.; Brulé-Babel, A. Mapping of Fhb2 on chromosome 6BS: A gene controlling Fusarium head blight field resistance in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2007, 114, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhang, X.; Hou, Y.; Cai, J.; Shen, X.; Zhou, T.; Xu, H.; Ohm, H.W.; Wang, H.; Li, A.; et al. High-density mapping of the major FHB resistance gene Fhb7 derived from Thinopyrum ponticum and its pyramiding with Fhb1 by marker-assisted selection. Theor. Appl. Genet. 2015, 128, 2301–2316. [Google Scholar] [CrossRef] [PubMed]
- Schnurbusch, T.; Paillard, S.; Fossati, D.; Messmer, M.; Schachermayr, G.; Winzeler, M.; Keller, B. Detection of QTLs for Stagonospora glume blotch resistance in Swiss winter wheat. Theor. Appl. Genet. 2003, 107, 1226–1234. [Google Scholar] [CrossRef] [PubMed]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Fedak, G.; Cao, W.; Wolfe, D.; Chi, D.; Xue, A. Molecular characterization of Fusarium resistance from Elymus repens introgressed into bread wheat. Cytol. Genet. 2017, 51, 130–133. [Google Scholar] [CrossRef]
- Kleijer, G.; Bronnimann, A.; Fossati, A. Chromosomal location of a dominant gene for resistance at the seedling stage to Septoria nodorum Berk. in the wheat variety’Atlas 66’. Zeitschrift fur Pflanzenzuchtung 1977, 78, 170–173. [Google Scholar]
- Murphy, N.E.; Loughman, R.; Wilson, R.; Lagudah, E.S.; Appels, R.; Jones, M.G. Resistance to Septoria nodorum blotch in the Aegilops tauschii accession RL5271 is controlled by a single gene. Euphytica 2000, 113, 227–231. [Google Scholar] [CrossRef]
- Ma, H.; Hughes, G.R. Genetic control and chromosomal location of Triticum timopheevii-derived resistance to Septoria nodorum blotch in durum wheat. Genome 1995, 38, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Tommasini, L.; Schnurbusch, T.; Fossati, D.; Mascher, F.; Keller, B. Association mapping of Stagonospora nodorum blotch resistance in modern European winter wheat varieties. Theor. Appl. Genet. 2007, 115, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Friesen, T.L.; Ling, H.; Meinhardt, S.W.; Oliver, R.P.; Rasmussen, J.B.; Faris, J.D. The Tsn1–ToxA interaction in the wheat–Stagonospora nodorum pathosystem parallels that of the wheat–tan spot system. Genome 2006, 49, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Friesen, T.L.; Chu, C.G.; Liu, Z.H.; Xu, S.S.; Halley, S.; Faris, J.D. Host-selective toxins produced by Stagonospora nodorum confer disease susceptibility in adult wheat plants under field conditions. Theor. Appl. Genet. 2009, 118, 1489–1497. [Google Scholar] [CrossRef] [PubMed]
- Patokar, C.; Sepsi, A.; Schwarzacher, T.; Kishii, M.; Heslop-Harrison, J.S. Molecular cytogenetic characterization of novel wheat-Thinopyrum bessarabicum recombinant lines carrying intercalary translocations. Chromosoma 2016, 125, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Makandar, R.; Nalam, V.J.; Lee, H.; Trick, H.N.; Dong, Y.; Shah, J. Salicylic acid regulates basal resistance to Fusarium head blight in wheat. Mol. Plant Microbe Interact. 2012, 25, 431–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.; Mackintosh, C.A.; Lewis, J.; Heinen, S.J.; Radmer, L.; Dill-Macky, R.; Baldridge, G.D.; Zeyen, R.J.; Muehlbauer, G.J. Transgenic wheat expressing a barley class II chitinase gene has enhanced resistance against Fusarium graminearum. J. Exp. Bot. 2008, 59, 2371–2378. [Google Scholar] [CrossRef] [PubMed]
- Volpi, C.; Janni, M.; Lionetti, V.; Bellincampi, D.; Favaron, F.; D’Ovidio, R. The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol. Plant Microbe Interact. 2011, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, Z.; Xu, H.; Zhou, M.; Du, L.; Zhang, Z. Overexpression of wheat lipid transfer protein gene TaLTP5 increases resistances to Cochliobolus sativus and Fusarium graminearum in transgenic wheat. Funct. Integr. Genom. 2012, 12, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Walters, D.R.; Ratsep, J.; Havis, N.D. Controlling crop diseases using induced resistance: Challenges for the future. J. Exp. Bot. 2013, 64, 1263–1280. [Google Scholar] [CrossRef] [PubMed]
- Pospieszny, H. Systemiczna odporność nabyta (Systemic Acquired Resistance-SAR) w integrowanej ochronie roślin. Prog. Plant Prot. 2016, 56, 436–442. [Google Scholar]
- Gholamnezhad, J.; Sanjarian, F.; Goltapeh, E.M.; Safaie, N.; Razavi, K. Effect of salicylic acid on enzyme activity in wheat in immediate early time after infection with Mycosphaerella graminicola. Sci. Agric. Bohem. 2016, 47, 1–8. [Google Scholar]
- Cuperlovic-Culf, M.; Wang, L.; Forseille, L.; Boyle, K.; Merkley, N.; Burton, I.; Fobert, P.R. Metabolic biomarker panels of response to Fusarium head blight infection in different wheat varieties. PLoS ONE 2016, 11, e0153642. [Google Scholar] [CrossRef] [PubMed]
- Motallebi, P.; Niknam, V.; Ebrahimzadeh, H.; Hashemi, M.; Enferadi, S.T. Exogenous methyl jasmonate treatment induces defense response against Fusarium culmorum in wheat seedlings. J. Plant Growth Regul. 2017, 36, 71–82. [Google Scholar] [CrossRef]
- Lewandowski, M.; Skoracka, A.; Szydło, W.; Kozak, M.; Druciarek, T.; Griffiths, D.A. Genetic and morphological diversity of Trisetacus species (Eriophyoidea: Phytoptidae) associated with coniferous trees in Poland: Phylogeny, barcoding, host and habitat specialization. Exp. Appl. Acarol. 2014, 63, 497–520. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Schierscher-Viret, B.; Fossati, D.; Brabant, C.; Schori, A.; Keller, B.; Krattinger, S.G. Unlocking the diversity of genebanks: Whole-genome marker analysis of Swiss bread wheat and spelt. Theor. Appl. Genet. 2018, 131, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Pan, Q.; He, F.; Akhunova, A.; Chao, S.; Trick, H.; Akhunov, E. Transgenerational CRISPR-Cas9 Activity Facilitates Multiplex Gene Editing in Allopolyploid Wheat. CRISPR J. 2018, 1, 65–74. [Google Scholar] [CrossRef]
- Puchta, H. Applying CRISPR/Cas for genome engineering in plants: The best is yet to come. Curr. Opin. Plant Biol. 2017, 36, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2015, 32, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.; Østergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 258. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K.; Kumar, J.; Alok, A.; Tuli, R. RNA-guided genome editing for target gene mutations in wheat. G3 Genes Genomes Genet. 2013, 3, 2233–2238. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947. [Google Scholar] [CrossRef] [PubMed]
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.; Konečná, E.; Belanto, J.J.; Starker, C.G.; MAthre, J.W.; Greenstein, R.L.; et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar] [PubMed]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014, 42, 10903–10914. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Akhunova, A.; Chao, S.; Akhunov, E. Optimizing multiplex CRISPR/Cas9-based genome editing for wheat. BioRxiv 2016. [Google Scholar] [CrossRef]
- Koch, A.; Kogel, K.H. Plant Gene Silencing: Mechanisms and Applications; Dalmay, T., Ed.; CABI, School of Biological Sciences, University of East Anglia: Norwich, UK, 2017; Chapter 9; Volume 5, p. 166. [Google Scholar]
- Kuck, K.; Stenzel, K.; Vors, J. Modern Crop Protection Compounds, 2nd ed.; Krämer, W., Schirmer, U., Jeschke, P., Witschel, M., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2012; Chapter 19; pp. 761–805. [Google Scholar]
- Koch, A.; Kumar, N.; Weber, L.; Keller, H.; Imani, J.; Kogel, K.H. Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase–encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. USA 2013, 110, 19324–19329. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef] [PubMed]


| Pathogen | Gene | Gene Expression Interaction Stage | Encoded Trait | Author |
|---|---|---|---|---|
| Z. tritici | Mg3LysM | Colonization | Suppression of defense responses in wheat in the first stage of infection | [23,24] |
| NEP1 | Formation of fruiting bodies | Necrotic factor | [19,23] | |
| ZtNIP1 | Formation of fruiting bodies | Necrotic factor | [19,23] | |
| Cellulase genes | Formation of fruiting bodies | Production of cellulase, a cell-wall degrading enzyme | [23,25] | |
| Xylanase genes | Formation of fruiting bodies | Production of xylanase, a cell-wall degrading enzyme | [23,25] | |
| Pectinase genes | Formation of fruiting bodies | Production of pectinase, a cell-wall degrading enzyme | [23,25] | |
| F. graminearum | Tri5 | Colonization | Deoxynivalenol (DON) synthesis | [26,27] |
| Mgv1 | Sexual reproduction | Fecundity, production of heterokaryons; formation of fungal cells; virulence | [28,29] | |
| Gpmk1 | Sexual reproduction | Formation of ascospores and perithecia; virulence | [28,29] | |
| Cps1 | No data | Production of enzyme CPS1 composed of two AMP-binding domains with an unknown biochemical faction, a potential virulence factor | [30] | |
| Fgl1 | Colonization | Production of lipase, a potential virulence factor | [31,32] | |
| Cellulase genes | Colonization/Penetration | Production of cellulase, a cell-wall degrading enzyme | [31,33] | |
| Xylanase genes | Colonization/Penetration | Production of xylanase, a cell-wall degrading enzyme | [31,33] | |
| P. nodorum | ToxA | Colonization | Necrotic factor | [34,35] |
| SNOG | Colonization | Family of genes encoding the synthesis of various phosphate transporters | [36] | |
| 5S ribosomal | During all stages of infection | Synthesis of 5S ribosomal subunits | [37] | |
| Cellulase genes | Colonization/Penetration | Production of cellulase, a cell-wall degrading enzyme | [36,38] | |
| Xylanase genes | Colonization/Penetration | Production of xylanase, a cell-wall degrading enzyme | [36,38] |
| Pathogen | Resistance Gene | Chromosome | Author |
|---|---|---|---|
| Z. tritici | Stb1 | 5BL | [108] |
| Stb18 | 6DS | [109] | |
| StbSm3 | 3AS | [106] | |
| StbWW | 1BS | [110] | |
| Stb6 | 3AS | [111] | |
| Stb16q | 3DL | [112] | |
| F. graminearum | Fhb1 | 3BS, 5AS | [113] |
| Fhb2 | 6BS | [113] | |
| Fhb3 | 7AL | [114] | |
| Fhb4 | 4BL | [114] | |
| Fhb5 | 5AS | [114] | |
| Fhb6 | 1AS | [114] | |
| P. nodorum | Qsng.sfr.3BS | 3BS | [115] |
| Qsnb.fcu-1A | 1A | [99] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duba, A.; Goriewa-Duba, K.; Wachowska, U. A Review of the Interactions between Wheat and Wheat Pathogens: Zymoseptoria tritici, Fusarium spp. and Parastagonospora nodorum. Int. J. Mol. Sci. 2018, 19, 1138. https://doi.org/10.3390/ijms19041138
Duba A, Goriewa-Duba K, Wachowska U. A Review of the Interactions between Wheat and Wheat Pathogens: Zymoseptoria tritici, Fusarium spp. and Parastagonospora nodorum. International Journal of Molecular Sciences. 2018; 19(4):1138. https://doi.org/10.3390/ijms19041138
Chicago/Turabian StyleDuba, Adrian, Klaudia Goriewa-Duba, and Urszula Wachowska. 2018. "A Review of the Interactions between Wheat and Wheat Pathogens: Zymoseptoria tritici, Fusarium spp. and Parastagonospora nodorum" International Journal of Molecular Sciences 19, no. 4: 1138. https://doi.org/10.3390/ijms19041138
APA StyleDuba, A., Goriewa-Duba, K., & Wachowska, U. (2018). A Review of the Interactions between Wheat and Wheat Pathogens: Zymoseptoria tritici, Fusarium spp. and Parastagonospora nodorum. International Journal of Molecular Sciences, 19(4), 1138. https://doi.org/10.3390/ijms19041138