MicroRNA Expression Analysis of In Vitro Dedifferentiated Human Pancreatic Islet Cells Reveals the Activation of the Pluripotency-Related MicroRNA Cluster miR-302s
Abstract
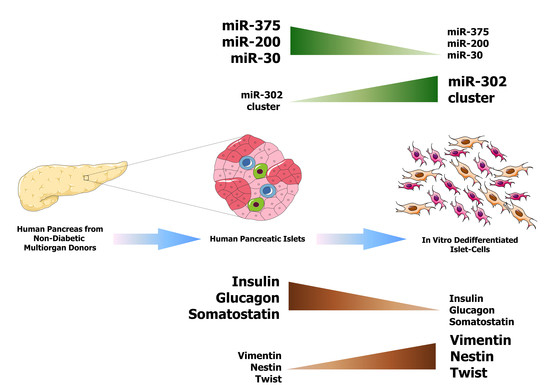
:1. Introduction
2. Results
2.1. MicroRNA Expression Profiles of In Vitro Dedifferentiated Islet Cells
2.2. miR-302 MicroRNAs Expression Is Switched-On during Islet Cells Dedifferentiation
2.3. Upregulated MicroRNA Target Key Genes with Multiple Roles in Endocrine/Epithelial Phenotype Maintenance
2.4. miR-302 MicroRNA Target Several Genes Involved in β-Cell Function and Are Regulated by Cell–Cell Contacts during In Vitro Dedifferentiation of Human Islets
3. Discussion
4. Materials and Methods
4.1. Human Pancreatic Islets Isolation and In Vitro Islet Dedifferentiation
4.2. RNA Extraction and Quality Evaluation
4.3. MicroRNA Expression Profiles Using Taqman Array Cards
4.4. MicroRNAs Stem–Loop Single Assay RT-qPCR
4.5. Predicted Target Genes Bioinformatic Analysis
4.6. Genes Expression Analysis
4.7. Statistical Analysis
4.8. Ethics Statement
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gallo, R.; Gambelli, F.; Gava, B.; Sasdelli, F.; Tellone, V.; Masini, M.; Marchetti, P.; Dotta, F.; Sorrentino, V. Generation and expansion of multipotent mesenchymal progenitor cells from cultured human pancreatic islets. Cell Death Differ. 2007, 14, 1860–1871. [Google Scholar] [CrossRef] [PubMed]
- Gershengorn, M.C.; Hardikar, A.A.; Wei, C.; Geras-Raaka, E.; Marcus-Samuels, B.; Raaka, B.M. Epithelial-to-mesenchymal transition generates proliferative human islet precursor cells. Science 2004, 306, 2261–2264. [Google Scholar] [CrossRef] [PubMed]
- Gershengorn, M.C.; Geras-Raaka, E.; Hardikar, A.A.; Raaka, B.M. Are better islet cell precursors generated by epithelial-to-mesenchymal transition? Cell Cycle 2005, 4, 380–382. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Amador, J.L.; Téllez, N.; Marin, S.; Aloy-Reverté, C.; Semino, C.; Nacher, M.; Montanya, E. Epithelial to mesenchymal transition in human endocrine islet cells. PLoS ONE 2018, 13, e0191104. [Google Scholar] [CrossRef] [PubMed]
- Russ, H.A.; Bar, Y.; Ravassard, P.; Efrat, S. In vitro proliferation of cells derived from adult human beta-cells revealed by cell-lineage tracing. Diabetes 2008, 57, 1575–1583. [Google Scholar] [CrossRef] [PubMed]
- Russ, H.A.; Ravassard, P.; Kerr-Conte, J.; Pattou, F.; Efrat, S. Epithelial-mesenchymal transition in cells expanded in vitro from lineage-traced adult human pancreatic beta cells. PLoS ONE 2009, 4, e6417. [Google Scholar] [CrossRef] [PubMed]
- Bar, Y.; Russ, H.A.; Sintov, E.; Anker-Kitai, L.; Knoller, S.; Efrat, S. Redifferentiation of expanded human pancreatic β-cell-derived cells by inhibition of the NOTCH pathway. J. Biol. Chem. 2012, 287, 17269–17280. [Google Scholar] [CrossRef] [PubMed]
- Grafi, G. The complexity of cellular dedifferentiation: Implications for regenerative medicine. Trends Biotechnol. 2009, 27, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Weir, G.C.; Aguayo-Mazzucato, C.; Bonner-Weir, S. β-cell dedifferentiation in diabetes is important, but what is it? Islets 2013, 5, 233–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weir, G.C.; Laybutt, D.R.; Kaneto, H.; Bonner-Weir, S.; Sharma, A. Beta-cell adaptation and decompensation during the progression of diabetes. Diabetes 2001, 50, S154–S159. [Google Scholar] [CrossRef] [PubMed]
- Cinti, F.; Bouchi, R.; Kim-Muller, J.Y.; Ohmura, Y.; Sandoval, P.R.; Masini, M.; Marselli, L.; Suleiman, M.; Ratner, L.E.; Marchetti, P.; et al. Evidence of β-Cell Dedifferentiation in Human Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2016, 101, 1044–1054. [Google Scholar] [CrossRef] [PubMed]
- Kim-Muller, J.Y.; Fan, J.; Kim, Y.J.R.; Lee, S.-A.; Ishida, E.; Blaner, W.S.; Accili, D. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic β cells in diabetic mice. Nat. Commun. 2016, 7, 12631. [Google Scholar] [CrossRef] [PubMed]
- Talchai, C.; Xuan, S.; Lin, H.V.; Sussel, L.; Accili, D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell 2012, 150, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Rui, J.; Deng, S.; Arazi, A.; Perdigoto, A.L.; Liu, Z.; Herold, K.C. β Cells that Resist Immunological Attack Develop during Progression of Autoimmune Diabetes in NOD Mice. Cell Metab. 2017, 25, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, P.; Bugliani, M.; de Tata, V.; Suleiman, M.; Marselli, L. Pancreatic beta cell identity in humans and the role of type 2 diabetes. Front. Cell Dev. Biol. 2017, 5, 55. [Google Scholar] [CrossRef] [PubMed]
- Marselli, L.; Suleiman, M.; Masini, M.; Campani, D.; Bugliani, M.; Syed, F.; Martino, L.; Focosi, D.; Scatena, F.; Olimpico, F.; et al. Are we overestimating the loss of beta cells in type 2 diabetes? Diabetologia 2014, 57, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Shenoy, A.; Blelloch, R.H. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat. Rev. Mol. Cell Biol. 2014, 15, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Li, M.A.; He, L. microRNAs as novel regulators of stem cell pluripotency and somatic cell reprogramming. Bioessays 2012, 34, 670–680. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, G.; Valentini, M.; Grieco, G.E.; Ventriglia, G.; Nigi, L.; Mancarella, F.; Pellegrini, S.; Martino, G.; Sordi, V.; Piemonti, L.; et al. MicroRNA expression profiles of human iPSCs differentiation into insulin-producing cells. Acta Diabetol. 2017, 54, 265–281. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, M.V.; Joglekar, V.M.; Hardikar, A.A. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr. Patterns 2009, 9, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Eliasson, L.; Esguerra, J.L.S. Role of non-coding RNAs in pancreatic beta-cell development and physiology. Acta Physiol. 2014, 211, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Nathan, G.; Kredo-Russo, S.; Geiger, T.; Lenz, A.; Kaspi, H.; Hornstein, E.; Efrat, S. MiR-375 promotes redifferentiation of adult human β cells expanded in vitro. PLoS ONE 2015, 10, e0122108. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, S. MiR-30 family and EMT in human fetal pancreatic islets. Islets 2009, 1, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zhu, X.; Dou, Y. The miR-302/367 cluster: A comprehensive update on its evolution and functions. Open Biol. 2015, 5, 150138. [Google Scholar] [CrossRef] [PubMed]
- Barroso-del Jesus, A.; Lucena-Aguilar, G.; Menendez, P. The miR-302-367 cluster as a potential stemness regulator in ESCs. Cell Cycle 2009, 8, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.D.; Venkatasubrahmanyam, S.; Jia, F.; Sun, N.; Butte, A.J.; Wu, J.C. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009, 18, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Barroso-delJesus, A.; Romero-López, C.; Lucena-Aguilar, G.; Melen, G.J.; Sanchez, L.; Ligero, G.; Berzal-Herranz, A.; Menendez, P. Embryonic stem cell-specific miR302-367 cluster: Human gene structure and functional characterization of its core promoter. Mol. Cell. Biol. 2008, 28, 6609–6619. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Deng, S.; Zhao, Z.; Zhang, H.; Xiao, J.; Song, W.; Gao, F.; Guan, Y. Oct4 regulates the miR-302 cluster in P19 mouse embryonic carcinoma cells. Mol. Biol. Rep. 2011, 38, 2155–2160. [Google Scholar] [CrossRef] [PubMed]
- Card, D.A.G.; Hebbar, P.B.; Li, L.; Trotter, K.W.; Komatsu, Y.; Mishina, Y.; Archer, T.K. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 2008, 28, 6426–6438. [Google Scholar] [CrossRef] [PubMed]
- Bräutigam, C.; Raggioli, A.; Winter, J. The Wnt/β-catenin pathway regulates the expression of the miR-302 cluster in mouse ESCs and P19 cells. PLoS ONE 2013, 8, e75315. [Google Scholar] [CrossRef] [PubMed]
- Ikonomou, L.; Geras-Raaka, E.; Raaka, B.M.; Gershengorn, M.C. β-catenin signalling in mesenchymal islet-derived precursor cells. Cell Prolif. 2008, 41, 474–491. [Google Scholar] [CrossRef] [PubMed]
- Lenz, A.; Toren-Haritan, G.; Efrat, S. Redifferentiation of adult human β cells expanded in vitro by inhibition of the WNT pathway. PLoS ONE 2014, 9, e112914. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.; Deroo, T.; Fujita, Y.; Itasaki, N. A positive role of cadherin in Wnt/β-catenin signalling during epithelial-mesenchymal transition. PLoS ONE 2011, 6, e23899. [Google Scholar] [CrossRef] [PubMed]
- Russ, H.A.; Sintov, E.; Anker-Kitai, L.; Friedman, O.; Lenz, A.; Toren, G.; Farhy, C.; Pasmanik-Chor, M.; Oron-Karni, V.; Ravassard, P.; et al. Insulin-producing cells generated from dedifferentiated human pancreatic beta cells expanded in vitro. PLoS ONE 2011, 6, e25566. [Google Scholar] [CrossRef] [PubMed]
- Accili, D.; Talchai, S.C.; Kim-Muller, J.Y.; Cinti, F.; Ishida, E.; Ordelheide, A.M.; Kuo, T.; Fan, J.; Son, J. When β-cells fail: Lessons from dedifferentiation. Diabetes Obes. Metab. 2016, 18, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Roefs, M.M.; Carlotti, F.; Jones, K.; Wills, H.; Hamilton, A.; Verschoor, M.; Durkin, J.M.W.; Garcia-Perez, L.; Brereton, M.F.; McCulloch, L.; et al. Increased vimentin in human α- and β-cells in type 2 diabetes. J. Endocrinol. 2017, 233, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Pullen, T.J.; Rutter, G.A. When less is more: The forbidden fruits of gene repression in the adult β-cell. Diabetes Obes. Metab. 2013, 15, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Sanchez, A.; Nguyen-Tu, M.-S.; Rutter, G.A. DICER Inactivation Identifies Pancreatic β-Cell “Disallowed” Genes Targeted by MicroRNAs. Mol. Endocrinol. 2015, 29, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Guay, C.; Regazzi, R. New emerging tasks for microRNAs in the control of β-cell activities. Biochim. Biophys. Acta 2016, 1861, 2121–2129. [Google Scholar] [CrossRef] [PubMed]
- Tattikota, S.G.; Rathjen, T.; McAnulty, S.J.; Wessels, H.-H.; Akerman, I.; van de Bunt, M.; Hausser, J.; Esguerra, J.L.S.; Musahl, A.; Pandey, A.K.; et al. Argonaute2 mediates compensatory expansion of the pancreatic β cell. Cell Metab. 2014, 19, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Nesca, V.; Guay, C.; Jacovetti, C.; Menoud, V.; Peyot, M.-L.; Laybutt, D.R.; Prentki, M.; Regazzi, R. Identification of particular groups of microRNAs that positively or negatively impact on beta cell function in obese models of type 2 diabetes. Diabetologia 2013, 56, 2203–2212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turcatel, G.; Rubin, N.; El-Hashash, A.; Warburton, D. MIR-99a and MIR-99b modulate TGF-β induced epithelial to mesenchymal plasticity in normal murine mammary gland cells. PLoS ONE 2012, 7, e31032. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Sun, Y.; Yuan, Y.; Han, Z.; Zhang, P.; Zhang, J.; You, M.J.; Teruya-Feldstein, J.; Wang, M.; Gupta, S.; et al. miR-100 induces epithelial-mesenchymal transition but suppresses tumorigenesis, migration and invasion. PLoS Genet. 2014, 10, e1004177. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Wang, Z.; Chen, J.; Xiang, T.; Li, Q.; Diao, X.; Zhu, B. MicroRNA-214 promotes epithelial-mesenchymal transition and metastasis in lung adenocarcinoma by targeting the suppressor-of-fused protein (Sufu). Oncotarget 2015, 6, 38705–38718. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Sun, Y.; He, P. MicroRNA-137 inhibits BMP7 to enhance the epithelial-mesenchymal transition of breast cancer cells. Oncotarget 2017, 8, 18348–18358. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.-C.; Wu, P.-H.; Huang, H.-C.; Chung, H.-Y.; Chou, Y.-C.; Cai, B.-H.; Kannagi, R. Downregulation of miR-199a/b-5p is associated with GCNT2 induction upon epithelial-mesenchymal transition in colon cancer. FEBS Lett. 2017, 591, 1902–1917. [Google Scholar] [CrossRef] [PubMed]
- Song, X.-F.; Wang, Q.-H.; Huo, R. Effects of microRNA-708 on Epithelial-Mesenchymal Transition, Cell Proliferation and Apoptosis in Melanoma Cells by Targeting LEF1 through the Wnt Signaling Pathway. Pathol. Oncol. Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Comas, J.; Moreno-Asso, A.; Moreno-Vedia, J.; Martín, M.; Castaño, C.; Marzà-Florensa, A.; Bofill-De Ros, X.; Mir-Coll, J.; Montané, J.; Fillat, C.; et al. Stress-Induced MicroRNA-708 Impairs β-Cell Function and Growth. Diabetes 2017, 66, 3029–3040. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, E.; Clynes, M.; Jeppesen, P.B.; O’Driscoll, L. Identification of microRNAs with a role in glucose stimulated insulin secretion by expression profiling of MIN6 cells. Biochem. Biophys. Res. Commun. 2010, 396, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.-Y.; Fang, W.; Lin, S.-L. The miR-302-Mediated Induction of Pluripotent Stem Cells (iPSC): Multiple Synergistic Reprogramming Mechanisms. Methods Mol. Biol. 2018, 1733, 283–304. [Google Scholar] [PubMed]







| Category | Term | Count | p Value |
|---|---|---|---|
| GOTERM_BP_FAT | GO:0007155 cell adhesion | 41 | 5.18 × 10−26 |
| GOTERM_BP_FAT | GO:0051094 positive regulation of developmental process | 21 | 9.12 × 10−15 |
| GOTERM_BP_FAT | GO:0006928 cell motion | 19 | 8.66 × 10−9 |
| GOTERM_BP_FAT | GO:0007267 cell–cell signaling | 16 | 2.88 × 10−5 |
| GOTERM_BP_FAT | GO:0040008 regulation of growth | 14 | 1.16 × 10−6 |
| GOTERM_BP_FAT | GO:0016055 Wnt receptor signaling pathway | 11 | 4.45 × 10−8 |
| GOTERM_BP_FAT | GO:0008284 positive regulation of cell proliferation | 10 | 0.00329 |
| GOTERM_BP_FAT | GO:0051046 regulation of secretion | 6 | 0.016312 |
| GOTERM_BP_FAT | GO:0032925 regulation of activin receptor signaling pathway | 4 | 4.40 × 10−5 |
| GOTERM_BP_FAT | GO:0046324 regulation of glucose import | 3 | 0.024159 |
| GOTERM_BP_FAT | GO:0032868 response to insulin stimulus | 8 | 8.33 × 10−6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sebastiani, G.; Grieco, G.E.; Brusco, N.; Ventriglia, G.; Formichi, C.; Marselli, L.; Marchetti, P.; Dotta, F. MicroRNA Expression Analysis of In Vitro Dedifferentiated Human Pancreatic Islet Cells Reveals the Activation of the Pluripotency-Related MicroRNA Cluster miR-302s. Int. J. Mol. Sci. 2018, 19, 1170. https://doi.org/10.3390/ijms19041170
Sebastiani G, Grieco GE, Brusco N, Ventriglia G, Formichi C, Marselli L, Marchetti P, Dotta F. MicroRNA Expression Analysis of In Vitro Dedifferentiated Human Pancreatic Islet Cells Reveals the Activation of the Pluripotency-Related MicroRNA Cluster miR-302s. International Journal of Molecular Sciences. 2018; 19(4):1170. https://doi.org/10.3390/ijms19041170
Chicago/Turabian StyleSebastiani, Guido, Giuseppina Emanuela Grieco, Noemi Brusco, Giuliana Ventriglia, Caterina Formichi, Lorella Marselli, Piero Marchetti, and Francesco Dotta. 2018. "MicroRNA Expression Analysis of In Vitro Dedifferentiated Human Pancreatic Islet Cells Reveals the Activation of the Pluripotency-Related MicroRNA Cluster miR-302s" International Journal of Molecular Sciences 19, no. 4: 1170. https://doi.org/10.3390/ijms19041170
APA StyleSebastiani, G., Grieco, G. E., Brusco, N., Ventriglia, G., Formichi, C., Marselli, L., Marchetti, P., & Dotta, F. (2018). MicroRNA Expression Analysis of In Vitro Dedifferentiated Human Pancreatic Islet Cells Reveals the Activation of the Pluripotency-Related MicroRNA Cluster miR-302s. International Journal of Molecular Sciences, 19(4), 1170. https://doi.org/10.3390/ijms19041170