α-Ketoglutarate Promotes Pancreatic Progenitor-Like Cell Proliferation
Abstract
:1. Introduction
2. Results
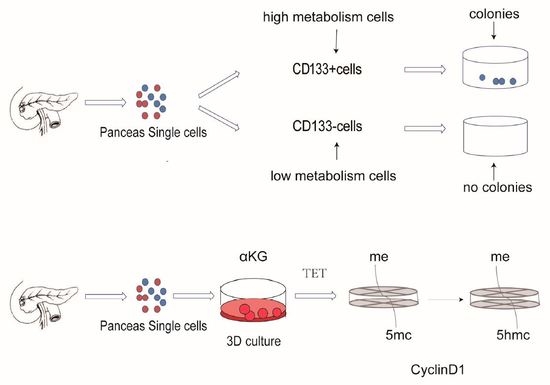
2.1. Metabolism Characterization of Pancreatic Progenitor-Like Cells from the Adult Mouse Pancreas
2.2. αKG Promoted the Proliferation of Pancreatic Progenitor-Like Cells
2.3. Succinate Suppresses the Proliferation of Pancreatic Progenitor-Like Cells
2.4. Regulation Proliferation of Pancreatic Progenitor-Like Cells via Tet
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Single Cell Preparation and 3D Culturing
4.3. RNA Extraction
4.4. Real-Time PCR and RT-PCR
4.5. Self-Renewal Assay
4.6. Immunostaining
4.7. Flow Cytometry and Cell Sorting
4.8. ATP Level Determination
4.9. Level of Reactive Oxide Species (ROS)
4.10. Quantification of Mitochondrial Number
4.11. Intermediate Metabolite Assay
4.12. Insulin Secretion Assay
4.13. Methylation-Specific PCR (MSP) Assay
4.14. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 3D culture system | three dimensional semisolid culture system |
| αKG | a-ketoglutarate |
| 5mc | 5-methylcytosine |
| 5hmc | 5-hydroxymethylcytosine |
| CK7 | cytokeratin |
| EGF | epidermal growth factor |
| FBS | fetal bovine serum |
| Nkx2.2 | Nk2 homeobox 2 |
| Nkx6.1 | Nk6 homeobox 1 |
| Pax4 | paired box 4 |
| Pax6 | paired box 6 |
| Pdx1 | pancreatic and duodenal homeobox 1 |
| Sox9 | (sex determining region Y)-box 9 |
| ROS | Reactive oxide species |
| MSP | methylation-specific PCR, |
| Oligo | oligomycin A |
| 2-DG | 2-Deoxy-d-glucose |
| Tet | tet methylcytosine dioxygenase |
| TCA | Tricaboxylic acid cycle |
| Cs | citrate synthase |
| Idh | isocitrate dehydrogenase |
| Ogdh | oxoglutarate (alpha-ketoglutarate) dehydrogenase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
Appendix A
High Throughput Sequencing
Appendix B
Appendix C
| Pathway Name | Significantly Enriched Genes | All Genes of Pathway | p-Value |
|---|---|---|---|
| Metabolic pathways | 543 | 1187 | 3.46 × 10−32 |
| Oxidative phosphorylation | 84 | 135 | 5.07 × 10−12 |
| Protein processing in endoplasmic reticulum | 92 | 162 | 5.33 × 10−10 |
| PI3K-Akt signaling pathway | 152 | 334 | 0.000000555 |
| Aminoacyl-tRNA biosynthesis | 28 | 41 | 0.00000586 |
| HIF-1 signaling pathway | 59 | 113 | 0.0000266 |
| Pyruvate metabolism | 25 | 41 | 0.000297 |
| MAPK signaling pathway | 109 | 252 | 0.000361 |
| Glycine, serine and threonine metabolism | 23 | 39 | 0.001 |
| Fatty acid metabolism | 26 | 46 | 0.00115 |
| Citrate cycle (TCA cycle) | 19 | 31 | 0.00146 |
| N-Glycan biosynthesis | 27 | 49 | 0.00155 |
| Notch signaling pathway | 26 | 49 | 0.00379 |
| Cysteine and methionine metabolism | 20 | 36 | 0.0055 |
| Amino sugar and nucleotide sugar metabolism | 25 | 48 | 0.00616 |
| Maturity onset diabetes of the young | 15 | 26 | 0.01 |
| Glutathione metabolism | 25 | 50 | 0.0118 |
| Fructose and mannose metabolism | 20 | 38 | 0.0119 |
| Pentose phosphate pathway | 16 | 29 | 0.0138 |
| Beta alanine metabolism | 16 | 29 | 0.0138 |
| Insulin signaling pathway | 58 | 137 | 0.0164 |
| Glyoxylate and dicarboxylate metabolism | 14 | 25 | 0.0178 |
| Alanine, aspartate and glutamate metabolism | 17 | 32 | 0.0178 |
| Purine metabolism | 67 | 165 | 0.0271 |
| Porphyrin and chlorophyll metabolism | 21 | 43 | 0.0273 |
| Sphingolipid metabolism | 20 | 41 | 0.0313 |
| Pyrimidine metabolism | 41 | 96 | 0.0356 |
| D-Glutamine and D-glutamate metabolism | 3 | 3 | 0.0383 |
| TGF-beta signaling pathway | 36 | 84 | 0.0451 |


Appendix D


References
- Mccall, M.; Shapiro, A.M. Islet cell transplantation. Semin. Pediatr. Surg. 2014, 23, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Huurman, V.A.; Hilbrands, R.; Pinkse, G.G.; Gillard, P.; Duinkerken, G.; Van de Linde, P.; van der Meer-Prins, P.M.; Versteeg-van der Voort, M.F.; Verbeeck, K.; Alizadeh, B.Z.; et al. Cellular Islet Autoimmunity Associates with Clinical Outcome of Islet Cell Transplantation. PLoS ONE 2008, 3, e2435. [Google Scholar] [CrossRef] [PubMed]
- Bonnerweir, S.; Sharma, A. Pancreatic Stem Cells; Humana Press: New York, NY, USA, 2009. [Google Scholar]
- Domínguez-Bendala, J. Pancreatic Stem Cells. J. Pathol. 2002, 197, 519–526. [Google Scholar]
- Stoffers, D.A.; Kieffer, T.J.; Hussain, M.A.; Drucker, D.J.; Bonner-Weir, S.; Habener, J.F.; Egan, J.M. Insulinotropic glucagon-like peptide 1 agonists stimulate expression of homeodomain protein IDX-1 and increase islet size.in mouse pancreas. Diabetes 2000, 49, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Oshima, Y.; Suzuki, A.; Kawashimo, K.; Ishikawa, M.; Ohkohchi, N.; Taniguchi, H. Isolation of mouse pancreatic ductal progenitor cells expressing CD133 and c-Met by flow cytometric cell sorting. Gastroenterology 2007, 132, 720–732. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Feng, T.; Shih, H.P.; Zerda, R.; Luo, A.; Hsu, J.; Mahdavi, A.; Sander, M.; Tirrell, D.A.; Riggs, A.D.; et al. Colony-forming cells in the adult mouse pancreas are expandable in Matrigel and form endocrine/acinar colonies in laminin hydrogel. Proc. Natl. Acad. Sci. USA 2013, 110, 3907–3912. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Feng, T.; Zerda, R.; Chen, C.C.; Riggs, A.D.; Ku, H.T. In vitro multilineage differentiation and self-renewal of single pancreatic colony-forming cells from adult C57BL/6 mice. Stem Cells Dev. 2014, 23, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Chung, I.; Lin, H.; Yang, D.; Liang, R.; Leung, A. Methionine aminopeptidase 2 (MetAP2) is required for zebrafish hematopoietic stem cell initiation. In Proceedings of the 7th Zebrafish European Conference (EZM 2011), Edinburgh, UK, 5–9 July 2011. [Google Scholar]
- Shiraki, N.; Shiraki, Y.; Tsuyama, T.; Obata, F.; Miura, M.; Nagae, G.; Aburatani, H.; Kume, K.; Endo, F.; Kume, S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014, 19, 780–794. [Google Scholar] [CrossRef] [PubMed]
- Dongshen, M.; Shanshan, T.; Jing, S. Culturing and Transcriptome Profiling of Progenitor like Colonies Derived from Adult Mouse Pancreas. Stem Cell Res. Ther. 2017, 8, 172–228. [Google Scholar]
- Shyhchang, N.; Daley, G.Q.; Cantley, L.C. Stem cell metabolism in tissue development and aging. Development 2013, 140, 2535–2547. [Google Scholar] [CrossRef] [PubMed]
- Rafalski, V.A.; Brunet, A. Energy metabolism in adult neural stem cell fate. Prog. Neurobiol. 2011, 93, 182–203. [Google Scholar] [CrossRef] [PubMed]
- Coloff, J.L.; Murphy, J.P.; Braun, C.R.; Harris, I.S.; Shelton, L.M.; Kami, K.; Gygi, S.P.; Selfors, L.M.; Brugge, J.S. Differential Glutamate Metabolism in Proliferating and Quiescent Mammary Epithelial Cells. Cell Metab. 2016, 23, 867–880. [Google Scholar] [CrossRef] [PubMed]
- TeSlaa, T.; Chaikovsky, A.C.; Lipchina, I.; Escobar, S.L.; Hochedlinger, K.; Huang, J.; Graeber, T.G.; Braas, D.; Teitell, M.A. α-Ketoglutarate Accelerates the Initial Differentiation of Primed Human Pluripotent Stem Cells. Cell Metab. 2016, 24, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Carey, B.W.; Finley, L.W.; Cross, J.R.; Allis, C.D.; Thompson, C.B. Intracellular a-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 2015, 518, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Berdasco, M.; Esteller, M. DNA methylation in stem cell renewal and multipotency. Stem Cell Res. Ther. 2011, 2, 42. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Krug, L.; Basu, A.; Meyer, P.; Treiber, N.; Vander Beken, S.; Wlaschek, M.; Kochanek, S.; Bloch, W.; Geiger, H.; et al. Alpha-Ketoglutarate Curbs Differentiation and Induces Cell Death in Mesenchymal Stromal Precursors with Mitochondrial Dysfunction. Stem Cells 2017, 35, 1704–1718. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.Y.; Kwak, S.; Lee, S.; Kim, H.; Lee, S.E.; Kim, J.H.; Kim, Y.A.; Jeon, Y.K.; Chung, D.H.; Jin, X.; et al. Psat1-Dependent Fluctuations in α-Ketoglutarate Affect the Timing of ESC Differentiation. Cell Metab. 2016, 24, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Zhai, Y.; Li, N.; Wu, C.; Zhu, H.; Wei, Z.; Bai, C.; Li, G.; Hua, J. Modification of Tet1 and histone methylation dynamics in dairy goat male germline stem cells. Cell Prolif. 2016, 49, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Koh, K.P.; Yabuuchi, A.; Rao, S.; Huang, Y.; Cunniff, K.; Nardone, J.; Laiho, A.; Tahiliani, M.; Sommer, C.A.; Mostoslavsky, G.; et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 2011, 8, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D.; Nicholls, D.G. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011, 435, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Yang, H.; Xu, W.; Ma, S.; Lin, H.; Zhu, H.; Liu, L.; Liu, Y.; Yang, C.; Xu, Y.; et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012, 26, 1326–1338. [Google Scholar] [CrossRef] [PubMed]
- Dunn, S.J.; Martello, G.; Yordanov, B.; Emmott, S.; Smith, A.G. Defining an essential transcription factor program for naïve pluripotency. Science 2014, 344, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Hackett, J.A.; Surani, M.A. Regulatory principles of pluripotency: From the ground state up. Cell Stem Cell 2014, 15, 416–430. [Google Scholar] [CrossRef] [PubMed]
- Suda, T.; Takubo, K.; Semenza, G.L. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 2011, 9, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Maruyama, T.; Fan, G. The Naive State of Human Pluripotent Stem Cells: A Synthesis of Stem Cell and Preimplantation Embryo Transcriptome Analyses. Cell Stem Cell 2014, 15, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Gafni, O.; Weinberger, L.; Mansour, A.A.; Manor, Y.S.; Chomsky, E.; Ben-Yosef, D.; Kalma, Y.; Viukov, S.; Maza, I.; Zviran, A.; et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013, 504, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Arrell, D.K.; Faustino, R.S.; Terzic, A.; Dzeja, P.P. Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. J. Mol. Cell. Cardiol. 2010, 48, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, A.; Woo, T.L.; Choo, A.B.; Reuveny, S.; Oh, S.K. Investigations into the metabolism of two-dimensional colony and suspended microcarrier cultures of human embryonic stem cells in serum-free media. Stem Cells Dev. 2010, 19, 1781–1792. [Google Scholar] [CrossRef] [PubMed]
- Varum, S.; Rodrigues, A.S.; Moura, M.B.; Momcilovic, O.; Easley, C.A., IV; Ramalho-Santos, J.; Van Houten, B.; Schatten, G. Energy Metabolism in Human Pluripotent Stem Cells and Their Differentiated Counterparts. PLoS ONE 2011, 6, e20914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondoh, H.; Lleonart, M.E.; Gil, J.; Wang, J.; Degan, P.; Peters, G.; Martinez, D.; Carnero, A.; Beach, D. Glycolytic Enzymes Can Modulate Cellular Life Span. Cancer Res. 2005, 65, 177–185. [Google Scholar] [PubMed]
- Gu, W.; Gaeta, X.; Sahakyan, A.; Chan, A.B.; Hong, C.S.; Kim, R.; Braas, D.; Plath, K.; Lowry, W.E.; Christofk, H.R. Glycolytic Metabolism Plays a Functional Role in Regulating Human Pluripotent Stem Cell State. Cell Stem Cell 2016, 19, 476–490. [Google Scholar] [CrossRef] [PubMed]
- Clouthier, S.G.; Luo, M.; Davis, A.; Mcdermott, S.; Jiagge, E.; Gheordunescu, E.; Luther, T.; Spitz, D.R.; Wicha, M.S. (Eds.) Selectively targeting EMT and MET breast cancer stem cell states by inhibiting dysregulated glycolytic and antioxidant pathways. In Proceedings of the A Alfred Taubman Institute Medical Research Institute Symposium, Ann Arbor, MI, USA, 18 March 2014. [Google Scholar]
- Varum, S.; Momcilovic, O.; Castro, C.; Ben-Yehudah, A.; Ramalho-Santos, J.; Navara, C. Enhancement of human embryonic stem cell pluripotency through inhibition of the mitochondrial respiratory chain. Stem Cell Res. 2009, 3, 142–156. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, Y.; Bai, F.; Zhang, J.Y.; Ma, S.H.; Liu, J.; Xu, Z.D.; Zhu, H.G.; Ling, Z.Q.; Ye, D.; et al. Tumor development is associated with decrease of Tet gene expression and 5-methylcytosine hydroxylation. Oncogene 2013, 32, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Shi, Y.G. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development 2012, 139, 1895–1902. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010, 466, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner Tet1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Kohli, R.M.; Zhang, Y. Tet enzymes, TDG and the dynamics of DNA demethylation. Nature 2013, 502, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Wu, H.; Diep, D.; Yamaguchi, S.; D’Alessio, A.C.; Fung, H.L.; Zhang, K.; Zhang, Y. Genome-wide analysis reveals Tet- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 2013, 153, 692–706. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Jin, L.; Wang, X.; Luo, A.; Hu, J.; Zheng, X.; Tsark, W.M.; Riggs, A.D.; Ku, H.T.; Huang, W. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc. Natl. Acad. Sci. USA 2013, 110, 17892–17897. [Google Scholar] [CrossRef] [PubMed]
- Tsumura, A.; Hayakawa, T.; Kumaki, Y.; Takebayashi, S.I.; Sakaue, M.; Matsuoka, C.; Shimotohno, K.; Ishikawa, F.; Li, E.; Ueda, H.R.; et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells Devoted Mol. Cell. Mech. 2006, 11, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Lin, K.; Song, J.; Wang, Y. Effects of Tet-induced oxidation products of 5-methylcytosine on Dnmt1- and DNMT3a-mediated cytosine methylation. Mol. Biosyst. 2014, 10, 1749–1752. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.W.; Choi, J.; Lee, S.Y.; Sung, S.; Yoo, H.J.; Kang, M.J.; Cheong, H.; Son, J. Autophagy is required for PDAC glutamine metabolism. Sci. Rep. 2016, 6, 37594. [Google Scholar] [CrossRef] [PubMed]






| Primer Name | Sequence (5′ to 3′) | |
|---|---|---|
| 1 | CD133-F | CTCCCATCAGTGGATAGAGAACT |
| 2 | CD133-R | ATACCCCCTTTTGACGAGGCT |
| 3 | CK7-F | ACGGATGGGGCTAACTTACAA |
| 4 | CK7-R | AGTCCTCGATTTGCTCGAACT |
| 5 | CyclinD1-F | GCGTACCCTGACACCAATCTC |
| 6 | CyclinD1-R | CTCCTCTTCGCACTTCTGCTC |
| 7 | glucagon-F | TTACTTTGTGGCTGGATTGCTT |
| 8 | glucagon-R | AGTGGCGTTTGTCTTCATTCA |
| 9 | insulin1-F | CACTTCCTACCCCTGCTGG |
| 10 | insulin1-R | ACCACAAAGATGCTGTTTGACA |
| 11 | Nkx2.2-F | CCGGGCGGAGAAAGGTATG |
| 12 | Nkx2.2-R | CTGTAGGCGGAAAAGGGGA |
| 13 | Nkx6.1-F | CTGCACAGTATGGCCGAGATG |
| 14 | Nkx6.1-R | CCGGGTTATGTGAGCCCAA |
| 15 | Neurod1-F | ATGACCAAATCATACAGCGAGAG |
| 16 | Neurod1-R | TCTGCCTCGTGTTCCTCGT |
| 17 | Pdx1-F | CCCCAGTTTACAAGCTCGCT |
| 18 | Pdx1-R | CTCGGTTCCATTCGGGAAAGG |
| 19 | Sox9-F | GAGCCGGATCTGAAGAGGGA |
| 20 | Sox9-R | GCTTGACGTGTGGCTTGTTC |
| 21 | ngn3-F | AGTGCTCAGTTCCAATTCCAC |
| 22 | ngn3-R | CGGCTTCTTCGCTTTTTGCTG |
| 23 | Ki67-F | ATCATTGACCGCTCCTTTAGGT |
| 24 | Ki67-R | GCTCGCCTTGATGGTTCCT |
| 25 | Pax4-F | AGGGGGACTCTTTGTGAATGG |
| 26 | Pax4-R | ACCTGTGCGGTAGTAGCGT |
| 27 | Gck-F | TGAGCCGGATGCAGAAGGA |
| 28 | Gck-R | GCAACATCTTTACACTGGCCT |
| 29 | Pfk-F | GGAGGCGAGAACATCAAGCC |
| 30 | Pfk-R | CGGCCTTCCCTCGTAGTGA |
| 31 | Pk-F | GCCGCCTGGACATTGACTC |
| 32 | Pk-R | CCATGAGAGAAATTCAGCCGAG |
| 33 | Ogdh-F | GTTTCTTCAAACGTGGGGTTCT |
| 34 | Ogdh-R | GCATGATTCCAGGGGTCTCAAA |
| 35 | Cs-F | GGACAATTTTCCAACCAATCTGC |
| 36 | Cs-R | TCGGTTCATTCCCTCTGCATA |
| 37 | Idh-1-F | ATGCAAGGAGATGAAATGACACG |
| 38 | Idh-1-R | GCATCACGATTCTCTATGCCTAA |
| 39 | Ldha-F | CCGTTACCTGATGGGAGAGA |
| 40 | Ldha-R | GTAGGCACTGTCCACCACCT |
| 41 | Tet1F | ACACAGTGGTGCTAATGCAG |
| 42 | Tet1R | AGCATGAACGGGAGAATCGG |
| 43 | Tet2F | AGAGAAGACAATCGAGAAGTCGG |
| 44 | Tet2R | CCTTCCGTACTCCCAAACTCAT |
| 45 | Tet3F | TGCGATTGTGTCGAACAAATAGT |
| 46 | Tet3R | TCCATACCGATCCTCCATGAG |
| 47 | Dnmt1-F | AAGAATGGTGTTGTCTACCGAC |
| 48 | Dnmt1-R | CATCCAGGTTGCTCCCCTTG |
| 49 | Dnmt3a-F | GAGGGAACTGAGACCCCAC |
| 50 | Dnmt3a-R | CTGGAAGGTGAGTCTTGGCA |
| 51 | Dnmt3b-F | AGCGGGTATGAGGAGTGCAT |
| 52 | Dnmt3b-R | GGGAGCATCCTTCGTGTCTG |
| 53 | Methy-cyclinD1-F | GAGTTTGTACGAGAGTTTAGGGTTC |
| 54 | Methy-cyclinD1-R | AAAAATAAATACGTTTCCGAATACG |
| 55 | Unmethy-cyclinD1-F | GTTTGTATGAGAGTTTAGGGTTTGT |
| 56 | Unmethy-cyclinD1-R | AAATAAATACATTTCCAAATACACC |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Ma, D.; Xing, Y.; Tang, S.; Alahdal, M.; Guo, J.; Pan, Y.; Zhang, Y.; Shen, Y.; Wu, Q.; et al. α-Ketoglutarate Promotes Pancreatic Progenitor-Like Cell Proliferation. Int. J. Mol. Sci. 2018, 19, 943. https://doi.org/10.3390/ijms19040943
Song J, Ma D, Xing Y, Tang S, Alahdal M, Guo J, Pan Y, Zhang Y, Shen Y, Wu Q, et al. α-Ketoglutarate Promotes Pancreatic Progenitor-Like Cell Proliferation. International Journal of Molecular Sciences. 2018; 19(4):943. https://doi.org/10.3390/ijms19040943
Chicago/Turabian StyleSong, Jing, Dongshen Ma, Yun Xing, Shanshan Tang, Murad Alahdal, Jiamin Guo, Yi Pan, Yanfeng Zhang, Yumeng Shen, Qiong Wu, and et al. 2018. "α-Ketoglutarate Promotes Pancreatic Progenitor-Like Cell Proliferation" International Journal of Molecular Sciences 19, no. 4: 943. https://doi.org/10.3390/ijms19040943
APA StyleSong, J., Ma, D., Xing, Y., Tang, S., Alahdal, M., Guo, J., Pan, Y., Zhang, Y., Shen, Y., Wu, Q., Lu, Z., & Jin, L. (2018). α-Ketoglutarate Promotes Pancreatic Progenitor-Like Cell Proliferation. International Journal of Molecular Sciences, 19(4), 943. https://doi.org/10.3390/ijms19040943