The Interaction of the Metallo-Glycopeptide Anti-Tumour Drug Bleomycin with DNA
Abstract
:1. Introduction
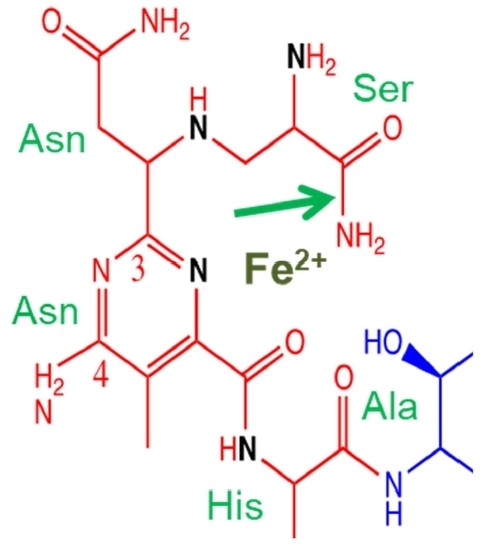
2. Bleomycin Is Composed of Four Functional Domains
3. Bleomycin DNA Cleavage Mechanism
4. Bleomycin Cleavage Specificity with Purified DNA
5. Bleomycin Cleavage Specificity with Purified DNA Using Updated Technology
6. Sequence Specificity at Bleomycin-Induced Abasic Sites
7. X-ray Crystal Structure of Bleomycin with DNA
8. Mechanism of Bleomycin-Induced Double-Strand Break Formation
9. The Sequence Specificity in Intact Human Cells
10. Sequence Specificity of Bleomycin Double-Strand Breaks in the Entire Human Genome
11. Comparison of the Bleomycin Genome-Wide DNA Sequence Specificity with Purified Plasmid DNA Sequences
12. Comparison of the Bleomycin Genome-Wide Sequence Specificity in Cellular DNA Compared with Purified Genomic DNA Sequences
13. Conformation of DNA and the DNA Sequence Specificity of Bleomycin
14. Chromatin Structure Affects the Interaction of Bleomycin with Cellular DNA
15. Cancer Signal Transduction Pathways Affected by Bleomycin
16. Repair of Bleomycin-Induced DNA Damage
16.1. Processing the 3′-Phosphoglycolate Termini
16.2. Genome-Wide Bleomycin Repair
16.3. Single-Strand Break Damage Repair
16.4. Repair of Double-Strand Breaks
16.5. Repair and Bleomycin Resistance
17. Cellular Transport of Bleomycin
18. Bleomycin Hydrolase
19. Bleomycin Analogues
20. Production of Novel Bleomycin Analogues That Are Resistant to Cleavage by Bleomycin Hydrolase
21. Summary and Future Prospects
- One approach is to produce bleomycin analogues that are resistant to cleavage by bleomycin hydrolase. The anti-tumour activity of bleomycin is limited by lung toxicity. The production of bleomycin analogues that are not cleaved by human bleomycin hydrolase will result in bleomycin analogues that are more effective as an anti-cancer agent because the lung toxicity would be eliminated.
- The engineering of an analogue that has improved uptake into cells, or even better, preferential uptake into tumour cells would produce a more effective cancer chemotherapeutic agent. Alterations to the disaccharide region could be prime areas for modification to achieve this aim.
- The creation of an analogue that is more efficient in producing the “activated” intermediate could have beneficial properties.
- More complicated and problematic would be the engineering of an analogue that is more effective at producing double-strand breaks compared with single-strand breaks, since double-strand breaks are thought to be the crucial lesion for the cytotoxicity of bleomycin.
- Further investigations with genome-wide studies will determine the crucial genes that are preferentially cleaved by bleomycin. In combination with nucleic acid-based techniques that target these crucial genes, bleomycin cytotoxicity could be enhanced by focusing on these important genes.
- Synergies could also be found with other nucleic acid and antibody-based novel therapies to enhance the action of these recently introduced therapeutics.
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| APE1 and APE2 | Apurinic/apyrimidinic endonuclease 1 and 2 |
| CE-LIF | Capillary electrophoresis with laser-induced fluorescence detection |
| TSS | Transcription start site |
| TDP1 | Tyrosyl-DNA phosphodiesterase 1 |
| ZBM | zorbamycin |
| M | A or C nucleotides |
| R | G or A nucleotides |
| W | A or T nucleotides |
| Y | T or C nucleotides |
References
- Umezawa, H.; Maeda, K.; Takeuchi, T.; Okami, Y. New antibiotics, bleomycin A and B. J. Antibiot. 1966, 19, 200–209. [Google Scholar] [PubMed]
- Du, L.; Sanchez, C.; Chen, M.; Edwards, D.J.; Shen, B. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem. Biol. 2000, 7, 623–642. [Google Scholar] [CrossRef]
- Shen, B.; Du, L.; Sanchez, C.; Edwards, D.J.; Chen, M.; Murrell, J.M. Cloning and characterization of the bleomycin biosynthetic gene cluster from Streptomyces verticillus ATCC15003. J. Nat. Prod. 2002, 65, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Du, L.; Sanchez, C.; Edwards, D.J.; Chen, M.; Murrell, J.M. The biosynthetic gene cluster for the anticancer drug bleomycin from Streptomyces verticillus ATCC15003 as a model for hybrid peptide-polyketide natural product biosynthesis. J. Ind. Microbiol. Biotechnol. 2001, 27, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Galm, U.; Wang, L.; Wendt-Pienkowski, E.; Yang, R.; Liu, W.; Tao, M.; Coughlin, J.M.; Shen, B. In vivo manipulation of the bleomycin biosynthetic gene cluster in Streptomyces verticillus ATCC15003 revealing new insights into its biosynthetic pathway. J. Biol. Chem. 2008, 283, 28236–28245. [Google Scholar] [CrossRef] [PubMed]
- Galm, U.; Wendt-Pienkowski, E.; Wang, L.; Huang, S.X.; Unsin, C.; Tao, M.; Coughlin, J.M.; Shen, B. Comparative analysis of the biosynthetic gene clusters and pathways for three structurally related antitumor antibiotics: Bleomycin, tallysomycin, and zorbamycin. J. Nat. Prod. 2011, 74, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, H. Studies on bleomycin. J. Formos Med. Assoc. 1969, 68, 569. [Google Scholar]
- Williams, S.D.; Birch, R.; Einhorn, L.H.; Irwin, L.; Greco, F.A.; Loehrer, P.J. Treatment of disseminated germ-cell tumors with cisplatin, bleomycin, and either vinblastine or etoposide. N. Engl. J. Med. 1987, 316, 1435–1440. [Google Scholar] [CrossRef] [PubMed]
- Stoter, G.; Kaye, S.B.; de Mulder, P.H.; Levi, J.; Raghavan, D. The importance of bleomycin in combination chemotherapy for good-prognosis germ cell carcinoma. J. Clin. Oncol. 1994, 12, 644–645. [Google Scholar] [CrossRef] [PubMed]
- Neese, F.; Zaleski, J.M.; Loeb Zaleski, K.; Solomon, E.I. Electronic Structure of Activated Bleomycin: Oxygen Intermediates in Heme versus Non-Heme Iron. J. Am. Chem. Soc. 2000, 122, 11703–11724. [Google Scholar] [CrossRef]
- Einhorn, L.H. Curing metastatic testicular cancer. Proc. Natl. Acad. Sci. USA 2002, 99, 4592–4595. [Google Scholar] [CrossRef] [PubMed]
- Froudarakis, M.; Hatzimichael, E.; Kyriazopoulou, L.; Lagos, K.; Pappas, P.; Tzakos, A.G.; Karavasilis, V.; Daliani, D.; Papandreou, C.; Briasoulis, E. Revisiting bleomycin from pathophysiology to safe clinical use. Crit. Rev. Oncol. Hematol. 2013, 87, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Carlson, R.W.; Sikic, B.I.; Turbow, M.M.; Ballon, S. Combination cisplatin, vinblastine, and bleomycin chemotherapy (PVB) for malignant germ-cell tumors of the ovary. J. Clin. Oncol. 1983, 1, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Leitheiser, C.J.; Smith, K.L.; Rishel, M.J.; Hashimoto, S.; Konishi, K.; Thomas, C.J.; Li, C.; McCormick, M.M.; Hecht, S.M. Solid-Phase Synthesis of Bleomycin Group Antibiotics. Construction of a 108-Member Deglycobleomycin Library. J. Am. Chem. Soc. 2003, 125, 8218–8227. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Stubbe, J. Bleomycins: Towards better therapeutics. Nat. Rev. Cancer 2005, 5, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Mirabelli, C.K.; Huang, C.H.; Fenwick, R.G.; Crooke, S.T. Quantitative measurement of single- and double-strand breakage of DNA in Escherichia coli by the antitumor antibiotics bleomycin and talisomycin. Antimicrob. Agents Chemother. 1985, 27, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Sikic, B.I. Biochemical and cellular determinants of bleomycin cytotoxicity. Cancer Surv. 1986, 5, 81–91. [Google Scholar] [PubMed]
- Povirk, L.F. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: Bleomycin, neocarzinostatin and other enediynes. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1996, 355, 71–89. [Google Scholar] [CrossRef]
- Tounekti, O.; Kenani, A.; Foray, N.; Orlowski, S.; Mir, L. The ratio of single-to double-strand DNA breaks and their absolute values determine cell death pathway. Br. J. Cancer 2001, 84, 1272. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, K.D.; Lewis, M.A.; Long, E.C.; Georgiadis, M.M. Crystal structure of DNA-bound Co(III) bleomycin B2: Insights on intercalation and minor groove binding. Proc. Natl. Acad. Sci. USA 2008, 105, 5052–5056. [Google Scholar] [CrossRef] [PubMed]
- Stubbe, J.; Kozarich, J.W.; Wu, W.; Vanderwall, D.E. Bleomycins: A structural model for specificity, binding, and double strand cleavage. Acc. Chem. Res. 1996, 29, 322–330. [Google Scholar] [CrossRef]
- Lehmann, T.; Topchiy, E. Contributions of NMR to the Understanding of the Coordination Chemistry and DNA Interactions of Metallo-Bleomycins. Molecules 2013, 18, 9253–9277. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Vanderwall, D.E.; Stubbe, J.; Kozarich, J.W.; Turner, C.J. Interaction of Co.cntdot.Bleomycin A2 (Green) with d(CCAGGCCTGG)2: Evidence for Intercalation Using 2D NMR. J. Am. Chem. Soc. 1994, 116, 10843–10844. [Google Scholar] [CrossRef]
- Deng, J.-Z.; Newman, D.J.; Hecht, S.M. Use of COMPARE analysis to discover functional analogues of bleomycin. J. Nat. Prod. 2000, 63, 1269–1272. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, J.; Sugiura, Y. Sequence-specific recognition and cleavage of DNA by metallobleomycin: Minor groove binding and possible interaction mode. Proc. Natl. Acad. Sci. USA 1988, 85, 2459–2463. [Google Scholar] [CrossRef] [PubMed]
- Manderville, R.A.; Ellena, J.F.; Hecht, S.M. Solution Structure of a Zn(II).Bleomycin A5-d(CGCTAGCG)2 Complex. J. Am. Chem. Soc. 1994, 116, 10851–10852. [Google Scholar] [CrossRef]
- Povirk, L.F.; Hogan, M.; Dattagupta, N. Binding of bleomycin to DNA: Intercalation of the bithiazole rings. Biochemistry 1979, 18, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Abraham, A.T.; Zhou, X.; Hecht, S.M. Metallobleomycin-Mediated Cleavage of DNA Not Involving a Threading-Intercalation Mechanism. J. Am. Chem. Soc. 2001, 123, 5167–5175. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.X.; Feng, Z.; Wang, L.; Galm, U.; Wendt-Pienkowski, E.; Yang, D.; Tao, M.; Coughlin, J.M.; Duan, Y.; Shen, B. A designer bleomycin with significantly improved DNA cleavage activity. J. Am. Chem. Soc. 2012, 134, 13501–13509. [Google Scholar] [CrossRef] [PubMed]
- Oppenheimer, N.J.; Chang, C.; Chang, L.H.; Ehrenfeld, G.; Rodriguez, L.O.; Hecht, S.M. Deglyco-bleomycin. Degradation of DNA and formation of a structurally unique Fe(II).CO complex. J. Biol. Chem. 1982, 257, 1606–1609. [Google Scholar] [PubMed]
- Chapuis, J.-C.; Schmaltz, R.M.; Tsosie, K.S.; Belohlavek, M.; Hecht, S.M. Carbohydrate Dependent Targeting of Cancer Cells by Bleomycin−Microbubble Conjugates. J. Am. Chem. Soc. 2009, 131, 2438–2439. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Paul, R.; Bhattacharya, C.; Bozeman, T.C.; Rishel, M.J.; Hecht, S.M. Structural Features Facilitating Tumor Cell Targeting and Internalization by Bleomycin and Its Disaccharide. Biochemistry 2015, 54, 3100–3109. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Schmaltz, R.M.; Bozeman, T.C.; Paul, R.; Rishel, M.J.; Tsosie, K.S.; Hecht, S.M. Selective tumor cell targeting by the disaccharide moiety of bleomycin. J. Am. Chem. Soc. 2013, 135, 2883–2886. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.R.; Ghare, M.I.; Bhattacharya, C.; Paul, R.; Yu, Z.; Zaleski, P.A.; Bozeman, T.C.; Rishel, M.J.; Hecht, S.M. The disaccharide moiety of bleomycin facilitates uptake by cancer cells. J. Am. Chem. Soc. 2014, 136, 13641–13656. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, C.; Yu, Z.; Rishel, M.J.; Hecht, S.M. The Carbamoylmannose Moiety of Bleomycin Mediates Selective Tumor Cell Targeting. Biochemistry 2014, 53, 3264–3266. [Google Scholar] [CrossRef] [PubMed]
- Madathil, M.M.; Bhattacharya, C.; Yu, Z.; Paul, R.; Rishel, M.J.; Hecht, S.M. Modified Bleomycin Disaccharides Exhibiting Improved Tumor Cell Targeting. Biochemistry 2014, 53, 6800–6810. [Google Scholar] [CrossRef] [PubMed]
- Dabrowiak, J.C. The coordination chemistry of bleomycin: A review. J. Biol. Inorg. Biochem. 1980, 13, 317–337. [Google Scholar] [CrossRef]
- Boger, D.L.; Ramsey, T.M.; Cai, H.; Hoehn, S.T.; Stubbe, J. Definition of the Effect and Role of the Bleomycin A 2 Valerate Substituents: Preorganization of a Rigid, Compact Conformation Implicated in Sequence-Selective DNA Cleavage. J. Am. Chem. Soc. 1998, 120, 9149–9158. [Google Scholar] [CrossRef]
- Boger, D.L.; Ramsey, T.M.; Cai, H.; Hoehn, S.T.; Stubbe, J. A Systematic Evaluation of the Bleomycin A 2 L -Threonine Side Chain: Its Role in Preorganization of a Compact Conformation Implicated in Sequence-Selective DNA Cleavage. J. Am. Chem. Soc. 1998, 120, 9139–9148. [Google Scholar] [CrossRef]
- Rishel, M.J.; Hecht, S.M. Analogues of bleomycin: Synthesis of conformationally rigid methylvalerates. Org. Lett. 2001, 3, 2867–2869. [Google Scholar] [CrossRef] [PubMed]
- Boger, D.L.; Teramoto, S.; Cai, H. N-methyl threonine analogues of deglycobleomycin A2: Synthesis and evaluation. Bioorg. Med. Chem. 1997, 5, 1577–1589. [Google Scholar] [CrossRef]
- Burger, R.M.; Peisach, J.; Horwitz, S.B. Activated bleomycin. A transient complex of drug, iron, and oxygen that degrades DNA. J. Biol. Chem. 1981, 256, 11636–11644. [Google Scholar] [PubMed]
- McLean, M.J.; Dar, A.; Waring, M.J. Differences between sites of binding to DNA and strand cleavage for complexes of bleomycin with iron of cobalt. J. Mol. Recognit. 1989, 1, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Stubbe, J.; Kozarich, J.W. Mechanisms of bleomycin-induced DNA degradation. Chem. Rev. 1987, 87, 1107–1136. [Google Scholar] [CrossRef]
- Chen, B.; Zhou, X.; Taghizadeh, K.; Chen, J.; Stubbe, J.; Dedon, P.C. GC/MS Methods To Quantify the 2-Deoxypentos-4-ulose and 3′-Phosphoglycolate Pathways of 4′ Oxidation of 2-Deoxyribose in DNA: Application to DNA Damage Produced by γ Radiation and Bleomycin. Chem. Res. Toxicol. 2007, 20, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Pitié, M.; Pratviel, G.V. Activation of DNA Carbon−Hydrogen Bonds by Metal Complexes. Chem. Rev. 2010, 110, 1018–1059. [Google Scholar] [CrossRef] [PubMed]
- Burger, R.M. Cleavage of Nucleic Acids by Bleomycin. Chem. Rev. 1998, 98, 1153–1170. [Google Scholar] [CrossRef] [PubMed]
- Magliozzo, R.S.; Peisach, J.; Ciriolo, M.R. Transfer RNA is cleaved by activated bleomycin. Mol. Pharmacol. 1989, 35, 428–432. [Google Scholar] [PubMed]
- Ekimoto, H.; Takahashi, K.; Matsuda, A.; Takita, T.; Umezawa, H. Lipid peroxidation by bleomycin-iron complexes in vitro. J. Antibiot. 1985, 38, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- Rana, T.M.; Meares, C.F. Transfer of oxygen from an artificial protease to peptide carbon during proteolysis. Proc. Natl. Acad. Sci. USA 1991, 88, 10578–10582. [Google Scholar] [CrossRef] [PubMed]
- Carter, B.J.; Murty, V.S.; Reddy, K.S.; Wang, S.N.; Hecht, S.M. A role for the metal binding domain in determining the DNA sequence selectivity of Fe-bleomycin. J. Biol. Chem. 1990, 265, 4193–4196. [Google Scholar] [PubMed]
- Morgan, M.A.; Hecht, S.M. Iron(II) Bleomycin-Mediated Degradation of a DNA-RNA Heteroduplex. Biochemistry 1994, 33, 10286–10293. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, M.; Grollman, A.P.; Ohtsubo, E.; Ohtsubo, H. Interaction of bleomycin with DNA. Proc. Natl. Acad. Sci. USA 1978, 75, 5983–5987. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, M.; Kappen, L.S.; Grollman, A.P.; Eisenberg, M.; Goldberg, I.H. Strand scission of deoxyribonucleic acid by neocarzinostatin, auromomycin, and bleomycin: Studies on base release and nucleotide sequence specificity. Biochemistry 1981, 20, 7599–7606. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, A.D.; Haseltine, W.A. Sequence specific cleavage of DNA by the antitumor antibiotics neocarzinostatin and bleomycin. Proc. Natl. Acad. Sci. USA 1978, 75, 3608–3612. [Google Scholar] [CrossRef] [PubMed]
- Kross, J.; Henner, W.D.; Hecht, S.M.; Haseltine, W.A. Specificity of deoxyribonucleic acid cleavage by bleomycin, phleomycin, and tallysomycin. Biochemistry 1982, 21, 4310–4318. [Google Scholar] [CrossRef] [PubMed]
- Mirabelli, C.K.; Ting, A.; Huang, C.H.; Mong, S.; Crooke, S.T. Bleomycin and talisomycin sequence-specific strand scission of DNA: A mechanism of double-strand cleavage. Cancer Res. 1982, 42, 2779–2785. [Google Scholar] [PubMed]
- Murray, V.; Martin, R.F. Comparison of the sequence specificity of bleomycin cleavage in two slightly different DNA sequences. Nucleic Acids Res. 1985, 13, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Murray, V.; Tan, L.; Matthews, J.; Martin, R.F. The sequence specificity of bleomycin damage in three cloned DNA sequences that differ by a small number of base substitutions. J. Biol. Chem. 1988, 263, 12854–12859. [Google Scholar] [PubMed]
- Nightingale, K.P.; Fox, K.R. DNA structure influences sequence specific cleavage by bleomycin. Nucleic Acids Res. 1993, 21, 2549–2555. [Google Scholar] [CrossRef] [PubMed]
- Murray, V. A survey of the sequence-specific interaction of damaging agents with DNA: Emphasis on anti-tumour agents. Prog. Nucleic Acid Res. Mol. Biol. 2000, 63, 367–415. [Google Scholar]
- Lewis, M.A.; Long, E.C. Fluorescent intercalator displacement analyses of DNA binding by the peptide-derived natural products netropsin, actinomycin, and bleomycin. Bioorg. Med. Chem. 2006, 14, 3481–3490. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Akiyama, Y.; Xu, Z.; Konishi, K.; Hecht, S.M. Identification and cleavage site analysis of DNA sequences bound strongly by bleomycin. J. Am. Chem. Soc. 2009, 131, 2013–2022. [Google Scholar] [CrossRef] [PubMed]
- Bozeman, T.C.; Nanjunda, R.; Tang, C.; Liu, Y.; Segerman, Z.J.; Zaleski, P.A.; Wilson, W.D.; Hecht, S.M. Dynamics of bleomycin interaction with a strongly bound hairpin DNA substrate, and implications for cleavage of the bound DNA. J. Am. Chem. Soc. 2012, 134, 17842–17845. [Google Scholar] [CrossRef] [PubMed]
- Segerman, Z.J.; Roy, B.; Hecht, S.M. Characterization of bleomycin-mediated cleavage of a hairpin DNA library. Biochemistry 2013, 52, 5315–5327. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Paul, A.; Alam, M.P.; Roy, B.; Wilson, W.D.; Hecht, S.M. A short DNA sequence confers strong bleomycin binding to hairpin DNAs. J. Am. Chem. Soc. 2014, 136, 13715–13726. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Tang, C.; Alam, M.P.; Hecht, S.M. DNA methylation reduces binding and cleavage by bleomycin. Biochemistry 2014, 53, 6103–6112. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Hecht, S.M. Hairpin DNA sequences bound strongly by bleomycin exhibit enhanced double-strand cleavage. J. Am. Chem. Soc. 2014, 136, 4382–4393. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, Y.; Ma, Q.; Edgar, E.; Laikhter, A.; Hecht, S.M. Identification of Strong DNA Binding Motifs for Bleomycin. J. Am. Chem. Soc. 2008, 130, 9650–9651. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, Y.; Ma, Q.; Edgar, E.; Laikhter, A.; Hecht, S.M. A Novel DNA Hairpin Substrate for Bleomycin. Org. Lett. 2008, 10, 2127–2130. [Google Scholar] [CrossRef] [PubMed]
- Giroux, R.A.; Hecht, S.M. Characterization of bleomycin cleavage sites in strongly bound hairpin DNAs. J. Am. Chem. Soc. 2010, 132, 16987–16996. [Google Scholar] [CrossRef] [PubMed]
- Murray, V.; Nguyen, T.V.; Chen, J.K. The Use of Automated Sequencing Techniques to Investigate the Sequence Selectivity of DNA Damaging Agents. Chem. Biol. Drug Des. 2012, 80, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Murray, V. Use of an Automated Capillary DNA Sequencer to Investigate the Interaction of Cisplatin with Telomeric DNA Sequences. Biomed. Chromatog. 2012, 26, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.Q.; Murray, V. The DNA sequence specificity of bleomycin cleavage in telomeric sequences in human cells. J. Biol. Inorg. Chem. 2012, 17, 1209–1215. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.V.; Murray, V. Human telomeric DNA sequences are a major target for the anti-tumour drug, bleomycin. J. Biol. Inorg. Chem. 2012, 17, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.V.; Chen, J.K.; Murray, V. Bleomycin DNA damage: Anomalous mobility of 3′-phosphoglycolate termini in an automated capillary DNA sequencer. J. Chromatogr. B 2013, 913, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.K.; Murray, V. The determination of the DNA sequence specificity of bleomycin-induced abasic sites. J. Biol. Inorg. Chem. 2016, 21, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Chung, L.H.; Murray, V. The mitochondrial DNA sequence specificity of the anti-tumour drug bleomycin using end-labeled DNA and capillary electrophoresis and a comparison with genome-wide DNA sequencing. J. Chromatogr. B 2016, 1008, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Gautam, S.D.; Chen, J.K.; Murray, V. The DNA sequence specificity of bleomycin cleavage in a systematically altered DNA sequence. J. Biol. Inorg. Chem. 2017, 22, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Murray, V.; Chen, J.K.; Tanaka, M.M. The genome-wide DNA sequence specificity of the anti-tumour drug bleomycin in human cells. Mol. Biol. Rep. 2016, 43, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.L. Hypoxia—A key regulatory factor in tumour growth. Nat. Rev. Cancer 2002, 2, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Wilson, W.R. Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer 2004, 4, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Povirk, L.F.; Wübker, W.; Köhnlein, W.; Hutchinson, F. DNA double-strand breaks and alkali-labile bonds produced by bleomycin. Nucleic Acids Res. 1977, 4, 3573–3580. [Google Scholar] [CrossRef] [PubMed]
- Povirk, L.F.; Han, Y.H.; Steighner, R.J. Structure of bleomycin-induced DNA double-strand breaks: Predominance of blunt ends and single-base 5′ extensions. Biochemistry 1989, 28, 5808–5814. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Mirabelli, C.K.; Jan, Y.; Crooke, S.T. Single-strand and double-strand deoxyribonucleic acid breaks produced by several bleomycin analogs. Biochemistry 1981, 20, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Absalon, M.J.; Kozarich, J.W.; Stubbe, J. Sequence Specific Double-Strand Cleavage of DNA by Fe-Bleomycin. 1. The Detection of Sequence-Specific Double-Strand Breaks Using Hairpin Oligonucleotides. Biochemistry 1995, 34, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Absalon, M.J.; Wu, W.; Kozarich, J.W.; Stubbe, J. Sequence-Specific Double-Strand Cleavage of DNA by Fe-Bleomycin. 2. Mechanism and Dynamics. Biochemistry 1995, 34, 2076–2086. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ghorai, M.K.; Kenney, G.; Stubbe, J. Mechanistic studies on bleomycin-mediated DNA damage: Multiple binding modes can result in double-stranded DNA cleavage. Nucleic Acids Res. 2008, 36, 3781–3790. [Google Scholar] [CrossRef] [PubMed]
- Steighner, R.J.; Povirk, L.F. Effect of in vitro cleavage of apurinic/apyrimidinic sites on bleomycin-induced mutagenesis of repackaged lambda phage. Mutat. Res. Genet. Toxicol. 1990, 240, 93–100. [Google Scholar] [CrossRef]
- Vanderwall, D.E.; Lui, S.M.; Wu, W.; Turner, C.J.; Kozarich, J.W.; Stubbe, J. A model of the structure of HOO-Co·bleomycin bound to d(CCAGTACTGG): Recognition at the d(GpT) site and implications for double-stranded DNA cleavage. Chem. Biol. 1997, 4, 373–387. [Google Scholar] [CrossRef]
- Boger, D.L.; Honda, T.; Dang, Q. Total synthesis of bleomycin A2 and related agents. 2. Synthesis of (−)-pyrimidoblamic acid, epi-(+)-pyrimidoblamic acid,(+)-desacetamidopyrimidoblamic acid, and (−)-descarboxamidopyrimidoblamic acid. J. Am. Chem. Soc. 1994, 116, 5619–5630. [Google Scholar] [CrossRef]
- Boger, D.L.; Colletti, S.L.; Teramoto, S.; Ramsey, T.M.; Zhou, J. Synthesis of key analogs of bleomycin A2 that permit a systematic evaluation of the linker region: Identification of an exceptionally prominent role for the L-threonine substituent. Bioorg. Med. Chem. 1995, 3, 1281–1295. [Google Scholar] [CrossRef]
- Murray, V.; Martin, R.F. The sequence specificity of bleomycin-induced DNA damage in intact cells. J. Biol. Chem. 1985, 260, 10389–10391. [Google Scholar] [PubMed]
- Cairns, M.J.; Murray, V. Influence of chromatin structure on bleomycin-DNA interactions at base pair resolution in the human beta-globin gene cluster. Biochemistry 1996, 35, 8753–8760. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Murray, V. A large “footprint” at the boundary of the human beta-globin locus control region hypersensitive site-2. Int. J. Biochem. Cell Biol. 2000, 32, 695–702. [Google Scholar] [CrossRef]
- Kim, A.; Murray, V. Chromatin structure at the 3′-boundary of the human beta-globin locus control region hypersensitive site-2. Int. J. Biochem. Cell Biol. 2001, 33, 1183–1192. [Google Scholar] [CrossRef]
- Temple, M.D.; Murray, V. Footprinting the ‘essential regulatory region’ of the retinoblatoma gene promoter in intact human cells. Int. J. Biochem. Cell Biol. 2005, 37, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Galea, A.M.; Murray, V. The influence of chromatin structure on DNA damage induced by nitrogen mustards and cisplatin analogues. Chem. Biol. Drug Des. 2010, 75, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Murray, V.; Chen, J.K.; Galea, A.M. The anti-tumour drug, bleomycin, preferentially cleaves at the transcription start sites of actively transcribed genes in human cells. Cell. Mol. Life Sci. 2014, 71, 1505–1512. [Google Scholar] [CrossRef] [PubMed]
- Murray, V.; Chen, J.K.; Galea, A.M. Enhanced repair of bleomycin DNA damage at the transcription start sites of actively transcribed genes in human cells. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2014, 769, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.K.; Yang, D.; Shen, B.; Murray, V. Bleomycin analogues preferentially cleave at the transcription start sites of actively transcribed genes in human cells. Int. J. Biochem. Cell Biol. 2017, 85, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Bolzán, A.D.; Bianchi, M.S. DNA and chromosome damage induced by bleomycin in mammalian cells: An update. Mutat. Res. Rev. Mutat. Res. 2018, 775, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Calladine, C.R. Mechanics of sequence-dependent stacking of bases in B-DNA. J. Mol. Biol. 1982, 161, 343–352. [Google Scholar] [CrossRef]
- Zgarbova, M.; Jurecka, P.; Lankas, F.; Cheatham, T.E., 3rd; Sponer, J.; Otyepka, M. Influence of BII Backbone Substates on DNA Twist: A Unified View and Comparison of Simulation and Experiment for All 136 Distinct Tetranucleotide Sequences. J. Chem. Inf. Model. 2017, 57, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Travers, A.A. The structural basis of DNA flexibility. Philos. Trans. A Math. Phys. Eng. Sci. 2004, 362, 1423–1438. [Google Scholar] [CrossRef] [PubMed]
- Geggier, S.; Vologodskii, A. Sequence dependence of DNA bending rigidity. Proc. Natl. Acad. Sci. USA 2010, 107, 15421–15426. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.T.; Hsu, T.C. Bleomycin causes release of nucleosomes from chromatin and chromosomes. Nature 1978, 271, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.T. Preferential damage of active chromatin by bleomycin. Cancer Res. 1981, 41, 2439–2443. [Google Scholar] [PubMed]
- Jiang, C.; Pugh, B.F. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009, 10, R109. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Pugh, B.F. Nucleosome positioning and gene regulation: Advances through genomics. Nat. Rev. Genet. 2009, 10, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Schones, D.E.; Cui, K.; Cuddapah, S.; Roh, T.Y.; Barski, A.; Wang, Z.; Wei, G.; Zhao, K. Dynamic regulation of nucleosome positioning in the human genome. Cell 2008, 132, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.D.; Fulmer, A.; Buckholz, J.; Zhang, B.; Cutrera, J.; Shiomitsu, K.; Li, S. Bleomycin/interleukin-12 electrochemogenetherapy for treating naturally occurring spontaneous neoplasms in dogs. Cancer Gene Ther. 2010, 17, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Guo, Y.; Huang, R.; Li, J.; Huang, S.; Kuang, Y.; Han, L.; Jiang, C. Gene and doxorubicin co-delivery system for targeting therapy of glioma. Biomaterials 2012, 33, 4907–4916. [Google Scholar] [CrossRef] [PubMed]
- Tounekti, O.; Pron, G.; Belehradek, J.; Mir, L.M. Bleomycin, an apoptosis-mimetic drug that induces two types of cell death depending on the number of molecules internalized. Cancer Res. 1993, 53, 5462–5469. [Google Scholar] [PubMed]
- Nelson, W.G.; Kastan, M.B. DNA strand breaks: The DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol. Cell. Biol. 1994, 14, 1815–1823. [Google Scholar] [CrossRef] [PubMed]
- Brahim, S.; Aroui, S.; Abid, K.; Kenani, A. Involvement of C-jun NH2-terminal kinase and apoptosis induced factor in apoptosis induced by deglycosylated bleomycin in laryngeal carcinoma cells. Cell Biol. Int. 2009, 33, 964–970. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.; Ensley, J.F.; Gutkind, J.S.; Yeudall, W.A. Induction of apoptosis in head-and-neck squamous carcinoma cells by gamma-irradiation and bleomycin is p53-independent. Int. J. Cancer 2000, 88, 737–743. [Google Scholar] [CrossRef]
- Gimonet, D.; Landais, E.; Bobichon, H.; Coninx, P.; Liautaud-Roger, F. Induction of apoptosis by bleomycin in p53-null HL-60 leukemia cells. Int. J. Oncol 2004, 24, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Niikura, Y.; Dixit, A.; Scott, R.; Perkins, G.; Kitagawa, K. BUB1 mediation of caspase-independent mitotic death determines cell fate. J. Cell Biol. 2007, 178, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, H.K.; Susin, S.A. Therapeutic potential of AIF-mediated caspase-independent programmed cell death. Drug Resist. Updates 2007, 10, 235–255. [Google Scholar] [CrossRef] [PubMed]
- Hardie, M.E.; Murray, V. The sequence preference of DNA cleavage by T4 Endonuclease VII. Biochimie 2018, 146, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Coulson, E.J. Does the p75 neurotrophin receptor mediate Abeta-induced toxicity in Alzheimer’s disease? J. Neurochem. 2006, 98, 654–660. [Google Scholar] [CrossRef] [PubMed]
- De Velasco, M.A.; Tanaka, M.; Yamamoto, Y.; Hatanaka, Y.; Koike, H.; Nishio, K.; Yoshikawa, K.; Uemura, H. Androgen deprivation induces phenotypic plasticity and promotes resistance to molecular targeted therapy in a PTEN-deficient mouse model of prostate cancer. Carcinogenesis 2014, 35, 2142–2153. [Google Scholar] [CrossRef] [PubMed]
- Boridy, S.; Le, P.U.; Petrecca, K.; Maysinger, D. Celastrol targets proteostasis and acts synergistically with a heat-shock protein 90 inhibitor to kill human glioblastoma cells. Cell Death Dis. 2014, 5, e1216. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.M. Processing of nonconventional DNA strand break ends. Environ. Mol. Mutagen. 2007, 48, 772–782. [Google Scholar] [CrossRef] [PubMed]
- Povirk, L.F. Processing of Damaged DNA Ends for Double-Strand Break Repair in Mammalian Cells. ISRN Mol. Biol. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Izumi, T.; Hazra, T.K.; Boldogh, I.; Tomkinson, A.E.; Park, M.S.; Ikeda, S.; Mitra, S. Requirement for human AP endonuclease 1 for repair of 3′-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis 2000, 21, 1329–1334. [Google Scholar] [CrossRef] [PubMed]
- Suh, D.; Wilson, D.M., 3rd; Povirk, L.F. 3′-phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 1997, 25, 2495–2500. [Google Scholar] [CrossRef] [PubMed]
- Burkovics, P.; Hajdú, I.; Szukacsov, V.; Unk, I.; Haracska, L. Role of PCNA-dependent stimulation of 3′-phosphodiesterase and 3′-5′ exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucleic Acids Res. 2009, 37, 4247–4255. [Google Scholar] [CrossRef] [PubMed]
- Burkovics, P.; Szukacsov, V.; Unk, I.; Haracska, L. Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 2006, 34, 2508–2515. [Google Scholar] [CrossRef] [PubMed]
- Moshous, D.; Callebaut, I.; de Chasseval, R.; Corneo, B.; Cavazzana-Calvo, M.; Le Deist, F.; Tezcan, I.; Sanal, O.; Bertrand, Y.; Philippe, N.; et al. Artemis, a Novel DNA Double-Strand Break Repair/V(D)J Recombination Protein, Is Mutated in Human Severe Combined Immune Deficiency. Cell 2001, 105, 177–186. [Google Scholar] [CrossRef]
- Ma, Y.; Pannicke, U.; Schwarz, K.; Lieber, M.R. Hairpin Opening and Overhang Processing by an Artemis/DNA-Dependent Protein Kinase Complex in Nonhomologous End Joining and V(D)J Recombination. Cell 2002, 108, 781–794. [Google Scholar] [CrossRef]
- Povirk, L.F.; Zhou, T.; Zhou, R.; Cowan, M.J.; Yannone, S.M. Processing of 3′-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J. Biol. Chem. 2007, 282, 3547–3558. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.; Kawahara, M.; Khan, I.S.; Yannone, S.M.; Povirk, L.F. Restoration of G1 chemo/radioresistance and double-strand-break repair proficiency by wild-type but not endonuclease-deficient Artemis. Nucleic Acids Res. 2011, 39, 6500–6510. [Google Scholar] [CrossRef] [PubMed]
- Inamdar, K.V.; Pouliot, J.J.; Zhou, T.; Lees-Miller, S.P.; Rasouli-Nia, A.; Povirk, L.F. Conversion of Phosphoglycolate to Phosphate Termini on 3′ Overhangs of DNA Double Strand Breaks by the Human Tyrosyl-DNA Phosphodiesterase hTdp1. J. Biol. Chem. 2002, 277, 27162–27168. [Google Scholar] [CrossRef] [PubMed]
- Raymond, A.C.; Staker, B.L.; Burgin, A.B. Substrate Specificity of Tyrosyl-DNA Phosphodiesterase I (Tdp1). J. Biol. Chem. 2005, 280, 22029–22035. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Akopiants, K.; Mohapatra, S.; Lin, P.-S.; Valerie, K.; Ramsden, D.A.; Lees-Miller, S.P.; Povirk, L.F. Tyrosyl-DNA phosphodiesterase and the repair of 3′-phosphoglycolate-terminated DNA double-strand breaks. DNA Repair 2009, 8, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Lee, J.W.; Tatavarthi, H.; Lupski, J.R.; Valerie, K.; Povirk, L.F. Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1). Nucleic Acids Res. 2005, 33, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Hirano, R.; Interthal, H.; Huang, C.; Nakamura, T.; Deguchi, K.; Choi, K.; Bhattacharjee, M.B.; Arimura, K.; Umehara, F.; Izumo, S.; et al. Spinocerebellar ataxia with axonal neuropathy: Consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007, 26, 4732–4743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murai, J.; Huang, S.Y.; Das, B.B.; Dexheimer, T.S.; Takeda, S.; Pommier, Y. Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells. J. Biol. Chem. 2012, 287, 12848–12857. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. DNA single-strand break repair. Exp. Cell Res. 2014, 329, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.J.; Kim, E.Y.; Demple, B. Excision of C-4′-oxidized deoxyribose lesions from double-stranded DNA by human apurinic/apyrimidinic endonuclease (Ape1 protein) and DNA polymerase beta. J. Biol. Chem. 1998, 273, 28837–28844. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, A.C.; Kreller, C.R.; Greenberg, M.M. Long Patch Base Excision Repair Compensates for DNA Polymerase β Inactivation by the C4′-Oxidized Abasic Site. Biochemistry 2011, 50, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. XRCC1 and DNA strand break repair. DNA Repair 2003, 2, 955–969. [Google Scholar] [CrossRef]
- Jasin, M.; Rothstein, R. Repair of Strand Breaks by Homologous Recombination. Cold Spring Harb. Perspect. Biol. 2013, 5, a012740. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.J.; Chen, D.J. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013, 2, 130–143. [Google Scholar] [PubMed]
- Robertson, K.A.; Bullock, H.A.; Xu, Y.; Tritt, R.; Zimmerman, E.; Ulbright, T.M.; Foster, R.S.; Einhorn, L.H.; Kelley, M.R. Altered expression of Ape1/ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Res. 2001, 61, 2220–2225. [Google Scholar] [PubMed]
- Alberts, D.S.; Chen, H.S.G.; Liu, R.; Himmelstein, K.J.; Mayersohn, M.; Perrier, D.; Gross, J.; Moon, T.; Broughton, A.; Salmon, S.E. Bleomycin pharmacokinetics in man. Cancer Chemother. Pharmacol. 1978, 1, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Dorr, R.T. Bleomycin pharmacology: Mechanism of action and resistance, and clinical pharmacokinetics. Semin. Oncol. 1992, 19, 3–8. [Google Scholar] [PubMed]
- Kanao, M.; Tomita, S.; Ishihara, S.; Murakami, A.; Okada, H. Chelation of bleomycin with copper in vivo. Chemotherapy 1973, 21, 1305–1310. [Google Scholar]
- Petering, D.H.; Byrnes, R.W.; Antholine, W.E. The role of redox-active metals in the mechanism of action of bleomycin. Chem. Biol. Interact. 1990, 73, 133–182. [Google Scholar] [CrossRef]
- Ehrenfeld, G.M.; Shipley, J.B.; Heimbrook, D.C.; Sugiyama, H.; Long, E.C.; van Boom, J.H.; van der Marel, G.A.; Oppenheimer, N.J.; Hecht, S.M. Copper-dependent cleavage of DNA by bleomycin. Biochemistry 1987, 26, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, Y.; Ishizu, K.; Miyoshi, K. Studies of metallobleomycins by electronic spectroscopy, electron spin resonance spectroscopy, and potentiometric titration. J. Antibiot. 1979, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Ehrenfeld, G.M.; Rodriguez, L.O.; Hecht, S.M.; Chang, C.; Basus, V.J.; Oppenheimer, N.J. Copper(I)-bleomycin: Structurally unique complex that mediates oxidative DNA strand scission. Biochemistry 1985, 24, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Pron, G.; Belehradek, J.; Mir, L.M. Identification of a Plasma Membrane Protein That Specifically Binds Bleomycin. Biochem. Biophys. Res. Commun. 1993, 194, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Pron, G.; Mahrour, N.; Orlowski, S.; Tounekti, O.; Poddevin, B.; Belehradek, J.; Mir, L.M. Internalisation of the bleomycin molecules responsible for bleomycin toxicity: A receptor-mediated endocytosis mechanism. Biochem. Pharmacol. 1999, 57, 45–56. [Google Scholar] [CrossRef]
- Aouida, M. A Genome-Wide Screen in Saccharomyces cerevisiae Reveals Altered Transport as a Mechanism of Resistance to the Anticancer Drug Bleomycin. Cancer Res. 2004, 64, 1102–1109. [Google Scholar] [CrossRef] [PubMed]
- Aouida, M.; Leduc, A.; Poulin, R.; Ramotar, D. AGP2 Encodes the Major Permease for High Affinity Polyamine Import in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 24267–24276. [Google Scholar] [CrossRef] [PubMed]
- Aouida, M.; Poulin, R.; Ramotar, D. The Human Carnitine Transporter SLC22A16 Mediates High Affinity Uptake of the Anticancer Polyamine Analogue Bleomycin-A5. J. Biol. Chem. 2010, 285, 6275–6284. [Google Scholar] [CrossRef] [PubMed]
- Berra, S.; Ayachi, S.; Ramotar, D. Upregulation of the Saccharomyces cerevisiae efflux pump Tpo1 rescues an Imp2 transcription factor-deficient mutant from bleomycin toxicity. Environ. Mol. Mutagen. 2014, 55, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Uemura, T.; Tachihara, K.; Tomitori, H.; Kashiwagi, K.; Igarashi, K. Characteristics of the polyamine transporter TPO1 and regulation of its activity and cellular localization by phosphorylation. J. Biol. Chem. 2005, 280, 9646–9652. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Kashiwagi, K. Characteristics of cellular polyamine transport in prokaryotes and eukaryotes. Plant. Physiol. Biochem. 2010, 48, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Della Latta, V.; Cecchettini, A.; Del Ry, S.; Morales, M. Bleomycin in the setting of lung fibrosis induction: From biological mechanisms to counteractions. Pharmacol. Res. 2015, 97, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Lazo, J.S.; Humphreys, C.J. Lack of metabolism as the biochemical basis of bleomycin-induced pulmonary toxicity. Proc. Natl. Acad. Sci. USA 1983, 80, 3064–3068. [Google Scholar] [CrossRef] [PubMed]
- Brömme, D.; Rossi, A.B.; Smeekens, S.P.; Anderson, D.C.; Payan, D.G. Human Bleomycin Hydrolase: Molecular Cloning, Sequencing, Functional Expression, and Enzymatic Characterization. Biochemistry 1996, 35, 6706–6714. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Fahmi, N.E.; Vialas, C.; Miller, G.M.; Hecht, S.M. Total Synthesis of Deamido Bleomycin A2, the Major Catabolite of the Antitumor Agent Bleomycin. J. Am. Chem. Soc. 2002, 124, 9476–9488. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.R.; Homanics, G.E.; Hoyt, D.G.; Klein, E.; Abernethy, J.; Lazo, J.S. The neutral cysteine protease bleomycin hydrolase is essential for epidermal integrity and bleomycin resistance. Proc. Natl. Acad. Sci. USA 1999, 96, 4680–4685. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.; Calmels, T.P.; Creutz, C.E.; Sebti, S.M. Yeast cysteine proteinase gene ycp1 induces resistance to bleomycin in mammalian cells. Mol. Pharmacol. 1995, 48, 676–681. [Google Scholar] [PubMed]
- Chen, J.; Chen, Y.; He, Q. Action of bleomycin is affected by bleomycin hydrolase but not by caveolin-1. Int. J. Oncol. 2012, 41, 2245–2252. [Google Scholar] [CrossRef] [PubMed]
- Okamura, Y.; Nomoto, S.; Hayashi, M.; Hishida, M.; Nishikawa, Y.; Yamada, S.; Fujii, T.; Sugimoto, H.; Takeda, S.; Kodera, Y. Identification of the bleomycin hydrolase gene as a methylated tumor suppressor gene in hepatocellular carcinoma using a novel triple-combination array method. Cancer Lett. 2011, 312, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ramotar, D. Cellular resistance to bleomycin in Saccharomyces cerevisiae is not affected by changes in bleomycin hydrolase levels. Biochem. Cell Biol. 2002, 80, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Xu, Z.; Schroeder, B.R.; Sun, W.; Wei, F.; Hashimoto, S.; Konishi, K.; Leitheiser, C.J.; Hecht, S.M. Biochemical evaluation of a 108-member deglycobleomycin library: Viability of a selection strategy for identifying bleomycin analogues with altered properties. J. Am. Chem. Soc. 2007, 129, 12439–12452. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.J.; Chizhov, A.O.; Leitheiser, C.J.; Rishel, M.J.; Konishi, K.; Tao, Z.F.; Hecht, S.M. Solid-phase synthesis of bleomycin A(5) and three monosaccharide analogues: Exploring the role of the carbohydrate moiety in RNA cleavage. J. Am. Chem. Soc. 2002, 124, 12926–12927. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.K.; Yang, D.; Shen, B.; Neilan, B.A.; Murray, V. Zorbamycin has a different DNA sequence selectivity compared with bleomycin and analogues. Bioorg. Med. Chem. 2016, 24, 6094–6101. [Google Scholar] [CrossRef] [PubMed]
- Murray, V.; Chen, J.K.; Yang, D.; Shen, B. The genome-wide sequence specificity of DNA cleavage by bleomycin analogues in human cells. 2018; manuscript submitted for publication. [Google Scholar]
- Yoshida, T.; Ogawa, M.; Ota, K.; Yoshida, Y.; Wakui, A.; Oguro, M.; Ariyoshi, Y.; Hirano, M.; Kimura, I.; Matsuda, T. Phase II study of NK313 in malignant lymphomas: An NK313 Malignant Lymphoma Study Group trial. Cancer Chemother. Pharmacol. 1993, 31, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Denny, W.A. DNA-Intercalating agents as antitumour drugs: Prospects for future design. Anticancer Drug Des. 1989, 4, 241–263. [Google Scholar] [PubMed]
- Murray, V.; Chen, J.K.; Galea, A.M. The Potential of Acridine Carboxamide Platinum complexes as Anti-Cancer Agents: A Review. Anti-Cancer Agents Med. Chem. 2014, 14, 695–705. [Google Scholar] [CrossRef]






| Study | Type of Break | Preferred Individual Nucleotides | Consensus Sequence from the Individual Nucleotide Data | Consensus Sequence from Complete Sequence Data | |||||
|---|---|---|---|---|---|---|---|---|---|
| Position | −3 | −2 | −1 | 0* | +1 | +2 | |||
| Early 32P-end-label experiments | SSB | T | G | T | 5′-TGT* | ||||
| Random DNA sequence | SSB | T | G | T | A | 5′-TGT*A | 5′-TGT*A | ||
| Systematically altered RTGTAY clone | SSB | C > T | C = T | G | T | A | T = A | 5′-YYGT*AW | |
| Purified DNA genome-wide preferred nucleotide (50k) | DSB | ns | T | G | T | A | T > A | 5′-TGT*AW | 5′-TGT*AT |
| Cellular DNA genome-wide preferred nucleotide (50k) | DSB | G | T | G | T | A | ns | 5′-GTGT*A | 5′-RTGT*AY |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murray, V.; Chen, J.K.; Chung, L.H. The Interaction of the Metallo-Glycopeptide Anti-Tumour Drug Bleomycin with DNA. Int. J. Mol. Sci. 2018, 19, 1372. https://doi.org/10.3390/ijms19051372
Murray V, Chen JK, Chung LH. The Interaction of the Metallo-Glycopeptide Anti-Tumour Drug Bleomycin with DNA. International Journal of Molecular Sciences. 2018; 19(5):1372. https://doi.org/10.3390/ijms19051372
Chicago/Turabian StyleMurray, Vincent, Jon K. Chen, and Long H. Chung. 2018. "The Interaction of the Metallo-Glycopeptide Anti-Tumour Drug Bleomycin with DNA" International Journal of Molecular Sciences 19, no. 5: 1372. https://doi.org/10.3390/ijms19051372
APA StyleMurray, V., Chen, J. K., & Chung, L. H. (2018). The Interaction of the Metallo-Glycopeptide Anti-Tumour Drug Bleomycin with DNA. International Journal of Molecular Sciences, 19(5), 1372. https://doi.org/10.3390/ijms19051372