Endogenous Purification of NR4A2 (Nurr1) Identified Poly(ADP-Ribose) Polymerase 1 as a Prime Coregulator in Human Adrenocortical H295R Cells
Abstract
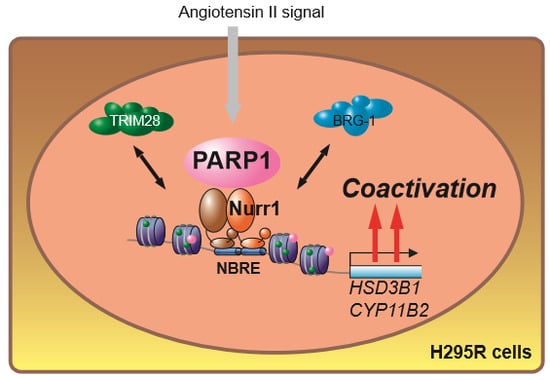
:1. Introduction
2. Results
2.1. Purification of Angiotensin II-Induced Nurr1-Associated Proteins in H295R Cells
2.2. Nurr1 Forms Stable Protein Complex with PARP1
2.3. PARP1 Supports Nurr1-Mediated Transcription Dependent on Enzymatic Activity
2.4. PARP1 Inhibitor Repressed Angiotensin II-Stimulated Aldosterone Secretion in H295R Cells
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Culture
4.3. RIME
4.4. In Vitro Translation and Pull down Assay
4.5. Western Blotting
4.6. Immunostaining
4.7. Luciferase Assay
4.8. RNA Isolation and Quantitative Real Time PCR
4.9. Measurement of Aldosterone Concentration
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PARP1 | poly(ADP-ribose) polymerase 1 |
| ZG | zona glomerulosa |
| MR | mineralocorticoid receptor |
| StAR | steroidgenic acute regulatory protein |
| DOC | deoxycorticosterone |
| PA | primary aldosteronism |
| TRH | treatment-resistant hypertension |
| RIME | rapid immunoprecipitation mass spectrometry of endogenous proteins |
| AngII | angiotensin II |
| IgG | immunogloblin G |
| DMSO | dimethyl sulfoxide |
| AMBIC | ammonium bicarbonate |
| NBRE | NGFI-B responsive element |
| PCR | polymerase chain reaction |
References
- Guagliardo, N.A.; Yao, J.; Hu, C.; Barrett, P.Q. Minireview: Aldosterone biosynthesis: Electrically gated for our protection. Endocrinology 2012, 153, 3579–3586. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Yamazaki, Y.; Konosu-Fukaya, S.; Ise, K.; Satoh, F.; Sasano, H. Aldosterone biosynthesis in the human adrenal cortex and associated disorders. J. Steroid Biochem. Mol. Biol. 2015, 153, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Funder, J. 30 years of the mineralocorticoid receptor: Mineralocorticoid receptor activation and specificity-conferring mechanisms: A brief history. J. Endocrinol. 2017, 234, T17–T21. [Google Scholar] [CrossRef] [PubMed]
- Connell, J.M.; MacKenzie, S.M.; Freel, E.M.; Fraser, R.; Davies, E. A lifetime of aldosterone excess: Long-term consequences of altered regulation of aldosterone production for cardiovascular function. Endocr. Rev. 2008, 29, 133–154. [Google Scholar] [CrossRef] [PubMed]
- Stocco, D.M. Tracking the role of a star in the sky of the new millennium. Mol. Endocrinol. 2001, 15, 1245–1254. [Google Scholar] [CrossRef] [PubMed]
- Connell, J.M.; Davies, E. The new biology of aldosterone. J. Endocrinol. 2005, 186, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Doi, M.; Satoh, F.; Maekawa, T.; Nakamura, Y.; Fustin, J.M.; Tainaka, M.; Hotta, Y.; Takahashi, Y.; Morimoto, R.; Takase, K.; et al. Isoform-specific monoclonal antibodies against 3β-hydroxysteroid dehydrogenase/isomerase family provide markers for subclassification of human primary aldosteronism. J. Clin. Endocrinol. Metab. 2014, 99, E257–E262. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Sanchez, C.E.; Lewis, M.; Nanba, K.; Rainey, W.E.; Kuppusamy, M.; Gomez-Sanchez, E.P. Development of monoclonal antibodies against the human 3β-hydroxysteroid dehydrogenase/isomerase isozymes. Steroids 2017, 127, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Mulatero, P.; Monticone, S.; Rainey, W.E.; Veglio, F.; Williams, T.A. Role of KCNJ5 in familial and sporadic primary aldosteronism. Nat. Rev. Endocrinol. 2013, 9, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Funder, J.W.; Carey, R.M.; Fardella, C.; Gomez-Sanchez, C.E.; Mantero, F.; Stowasser, M.; Young, W.F., Jr.; Montori, V.M. Case detection, diagnosis, and treatment of patients with primary aldosteronism: An endocrine society clinical practice guideline. J. Clin. Endocrinol. Metab. 2008, 93, 3266–3281. [Google Scholar] [CrossRef] [PubMed]
- Marzano, L.; Colussi, G.; Sechi, L.A.; Catena, C. Adrenalectomy is comparable with medical treatment for reduction of left ventricular mass in primary aldosteronism: Meta-analysis of long-term studies. Am. J. Hypertens. 2015, 28, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, H.K.; Menard, J.; White, W.B.; Young, W.F., Jr.; Williams, G.H.; Williams, B.; Ruilope, L.M.; McInnes, G.T.; Connell, J.M.; MacDonald, T.M. A double-blind, randomized study comparing the antihypertensive effect of eplerenone and spironolactone in patients with hypertension and evidence of primary aldosteronism. J. Hypertens. 2011, 29, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Eirin, A.; Textor, S.C.; Lerman, L.O. Emerging concepts for patients with treatment-resistant hypertension. Trends Cardiovasc. Med. 2016, 26, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Ito, R.; Sato, I.; Tsujita, T.; Yokoyama, A.; Sugawara, A. A ubiquitin-proteasome inhibitor bortezomib suppresses the expression of CYP11B2, a key enzyme of aldosterone synthesis. Biochem. Biophys. Res. Commun. 2017, 489, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Bassett, M.H.; Suzuki, T.; Sasano, H.; White, P.C.; Rainey, W.E. The orphan nuclear receptors NURR1 and NGFIB regulate adrenal aldosterone production. Mol. Endocrinol. 2004, 18, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, E.F.; Vargas, C.A.; Otis, M.; Gallo-Payet, N.; Bollag, W.B.; Rainey, W.E. Angiotensin-II acute regulation of rapid response genes in human, bovine, and rat adrenocortical cells. J. Mol. Endocrinol. 2007, 39, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.G.; Rilli, S.; Plonczynski, M.W.; Yanes, L.L.; Zhou, M.Y.; Gomez-Sanchez, E.P.; Gomez-Sanchez, C.E. Adrenal transcription regulatory genes modulated by angiotensin II and their role in steroidogenesis. Physiol. Genom. 2007, 30, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, M.A.; Muscat, G.E. The NR4A subgroup: Immediate early response genes with pleiotropic physiological roles. Nucl. Recept. Signal. 2006, 4, e002. [Google Scholar] [CrossRef] [PubMed]
- Kurakula, K.; Koenis, D.S.; van Tiel, C.M.; de Vries, C.J. NR4A nuclear receptors are orphans but not lonesome. Biochim. Biophys. Acta 2014, 1843, 2543–2555. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.; Takezawa, S.; Schule, R.; Kitagawa, H.; Kato, S. Transrepressive function of TLX requires the histone demethylase LSD1. Mol. Cell. Biol. 2008, 28, 3995–4003. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.; Katsura, S.; Ito, R.; Hashiba, W.; Sekine, H.; Fujiki, R.; Kato, S. Multiple post-translational modifications in hepatocyte nuclear factor 4α. Biochem. Biophys. Res. Commun. 2011, 410, 749–753. [Google Scholar] [CrossRef] [PubMed]
- Mullican, S.E.; Dispirito, J.R.; Lazar, M.A. The orphan nuclear receptors at their 25-year reunion. J. Mol. Endocrinol. 2013, 51, T115–T140. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, H.; Taylor, C.; Brown, G.D.; Papachristou, E.K.; Carroll, J.S.; D'Santos, C.S. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 2016, 11, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Maira, M.; Martens, C.; Philips, A.; Drouin, J. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol. Cell. Biol. 1999, 19, 7549–7557. [Google Scholar] [CrossRef] [PubMed]
- Rambaud, J.; Desroches, J.; Balsalobre, A.; Drouin, J. Tif1β/KAP-1 is a coactivator of the orphan nuclear receptor NGFI-B/Nur77. J. Biol. Chem. 2009, 284, 14147–14156. [Google Scholar] [CrossRef] [PubMed]
- Bilodeau, S.; Vallette-Kasic, S.; Gauthier, Y.; Figarella-Branger, D.; Brue, T.; Berthelet, F.; Lacroix, A.; Batista, D.; Stratakis, C.; Hanson, J.; et al. Role of Brg1 and HDAC2 in GR trans-repression of the pituitary pomc gene and misexpression in cushing disease. Genes Dev. 2006, 20, 2871–2886. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.A.; Kraus, W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liang, C.; Li, F.; Guan, D.; Wu, X.; Fu, X.; Lu, A.; Zhang, G. PARP1 in carcinomas and PARP1 inhibitors as antineoplastic drugs. Int. J. Mol. Sci. 2017, 18, 2111. [Google Scholar] [CrossRef] [PubMed]
- Ohkura, N.; Nagamura, Y.; Tsukada, T. Differential transactivation by orphan nuclear receptor NOR1 and its fusion gene product EWS/NOR1: Possible involvement of poly(ADP-ribose) polymerase I, PARP-1. J. Cell. Biochem. 2008, 105, 785–800. [Google Scholar] [CrossRef] [PubMed]
- Ota, T.; Doi, M.; Yamazaki, F.; Yarimizu, D.; Okada, K.; Murai, I.; Hayashi, H.; Kunisue, S.; Nakagawa, Y.; Okamura, H. Angiotensin II triggers expression of the adrenal gland zona glomerulosa-specific 3β-hydroxysteroid dehydrogenase isoenzyme through De Novo protein synthesis of the orphan nuclear receptors NGFIB and NURR1. Mol. Cell. Biol. 2014, 34, 3880–3894. [Google Scholar] [CrossRef] [PubMed]
- Curtin, N.J.; Wang, L.Z.; Yiakouvaki, A.; Kyle, S.; Arris, C.A.; Canan-Koch, S.; Webber, S.E.; Durkacz, B.W.; Calvert, H.A.; Hostomsky, Z.; et al. Novel poly(ADP-ribose) polymerase-1 inhibitor, AG14361, restores sensitivity to temozolomide in mismatch repair-deficient cells. Clin. Cancer Res. 2004, 10, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, E.; Hussein, M.A.; Savas, J.N.; Ouimet, M.; Barrett, T.J.; Leone, S.; Yates, J.R., 3rd; Moore, K.J.; Fisher, E.A.; Garabedian, M.J. Poly(ADP-ribose) polymerase 1 represses liver X receptor-mediated ABCA1 expression and cholesterol efflux in macrophages. J. Biol. Chem. 2016, 291, 11172–11184. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, F.; Wang, L.; Zhang, Y.; Li, X.; Huang, K.; Du, M.; Liu, F.; Huang, S.; Guan, Y.; et al. Poly(ADP-ribose) polymerase 1 promotes oxidative-stress-induced liver cell death via suppressing farnesoid X receptor α. Mol. Cell. Biol. 2013, 33, 4492–4503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, Y.; Wang, L.; Luo, X.; Huang, K.; Wang, C.; Du, M.; Liu, F.; Luo, T.; Huang, D. Poly(ADP-ribose) polymerase 1 is a key regulator of estrogen receptor α-dependent gene transcription. J. Biol. Chem. 2013, 288, 11348–11357. [Google Scholar] [CrossRef] [PubMed]
- Horsman, M.R. Nicotinamide and other benzamide analogs as agents for overcoming hypoxic cell radiation resistance in tumours. A review. Acta Oncol. 1995, 34, 571–587. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, C.R.; Almassy, R.; Barton, S.; Batey, M.A.; Calvert, A.H.; Canan-Koch, S.; Durkacz, B.W.; Hostomsky, Z.; Kumpf, R.A.; Kyle, S.; et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J. Natl. Cancer Inst. 2004, 96, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Kraus, W.L. Transcriptional control by PARP-1: Chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 2008, 20, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.A.; Zhang, Y.; Jiang, H.; Hussey, K.M.; Shrimp, J.H.; Lin, H.; Schwede, F.; Yu, Y.; Kraus, W.L. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 2016, 353, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.; Okuno, Y.; Chikanishi, T.; Hashiba, W.; Sekine, H.; Fujiki, R.; Kato, S. KIAA1718 is a histone demethylase that erases repressive histone methyl marks. Genes Cells 2010, 15, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.; Igarashi, K.; Sato, T.; Takagi, K.; Otsuka, I.M.; Shishido, Y.; Baba, T.; Ito, R.; Kanno, J.; Ohkawa, Y.; et al. Identification of myelin transcription factor 1 (MYT1) as a subunit of the neural cell type-specific lysine-specific demethylase 1 (LSD1) complex. J. Biol. Chem. 2014, 289, 18152–18162. [Google Scholar] [CrossRef] [PubMed]
- Shimada, H.; Kogure, N.; Noro, E.; Kudo, M.; Sugawara, K.; Sato, I.; Shimizu, K.; Kobayashi, M.; Suzuki, D.; Parvin, R.; et al. High glucose stimulates expression of aldosterone synthase (CYP11B2) and secretion of aldosterone in human adrenal cells. FEBS Open Bio 2017, 7, 1410–1421. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, D.; Saito-Hakoda, A.; Ito, R.; Shimizu, K.; Parvin, R.; Shimada, H.; Noro, E.; Suzuki, S.; Fujiwara, I.; Kagechika, H.; et al. Suppressive effects of RXR agonist PA024 on adrenal CYP11B2 expression, aldosterone secretion and blood pressure. PLoS ONE 2017, 12, e0181055. [Google Scholar] [CrossRef] [PubMed]





| Target Gene | Sequence |
| hGAPDH_F | atcccatcaccatcttccag |
| hGAPDH_R | atgagtccttccacgatacc |
| hCYP11B2_F | ggcagaggcagagatgctg |
| hCYP11B2_R | cttgagttagtgtctccaccagga |
| hCYP11B2_probe | (FAM) ctgcaccacgtgctgaagcact (TAM) |
| hHSD3B1_F | agaagagcctctggaaaacacatg |
| hHSD3B1_R | taaggcacaagtgtacagggtgc |
| hHSD3B1_probe | (FAM) ccatacccacacagc (TAM) |
| hPARP1_F | gctcctgaacaatgcagaca |
| hPARP1_R | cattgtgtgtggttgcatga |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noro, E.; Yokoyama, A.; Kobayashi, M.; Shimada, H.; Suzuki, S.; Hosokawa, M.; Takehara, T.; Parvin, R.; Shima, H.; Igarashi, K.; et al. Endogenous Purification of NR4A2 (Nurr1) Identified Poly(ADP-Ribose) Polymerase 1 as a Prime Coregulator in Human Adrenocortical H295R Cells. Int. J. Mol. Sci. 2018, 19, 1406. https://doi.org/10.3390/ijms19051406
Noro E, Yokoyama A, Kobayashi M, Shimada H, Suzuki S, Hosokawa M, Takehara T, Parvin R, Shima H, Igarashi K, et al. Endogenous Purification of NR4A2 (Nurr1) Identified Poly(ADP-Ribose) Polymerase 1 as a Prime Coregulator in Human Adrenocortical H295R Cells. International Journal of Molecular Sciences. 2018; 19(5):1406. https://doi.org/10.3390/ijms19051406
Chicago/Turabian StyleNoro, Erika, Atsushi Yokoyama, Makoto Kobayashi, Hiroki Shimada, Susumu Suzuki, Mari Hosokawa, Tomohiro Takehara, Rehana Parvin, Hiroki Shima, Kazuhiko Igarashi, and et al. 2018. "Endogenous Purification of NR4A2 (Nurr1) Identified Poly(ADP-Ribose) Polymerase 1 as a Prime Coregulator in Human Adrenocortical H295R Cells" International Journal of Molecular Sciences 19, no. 5: 1406. https://doi.org/10.3390/ijms19051406
APA StyleNoro, E., Yokoyama, A., Kobayashi, M., Shimada, H., Suzuki, S., Hosokawa, M., Takehara, T., Parvin, R., Shima, H., Igarashi, K., & Sugawara, A. (2018). Endogenous Purification of NR4A2 (Nurr1) Identified Poly(ADP-Ribose) Polymerase 1 as a Prime Coregulator in Human Adrenocortical H295R Cells. International Journal of Molecular Sciences, 19(5), 1406. https://doi.org/10.3390/ijms19051406