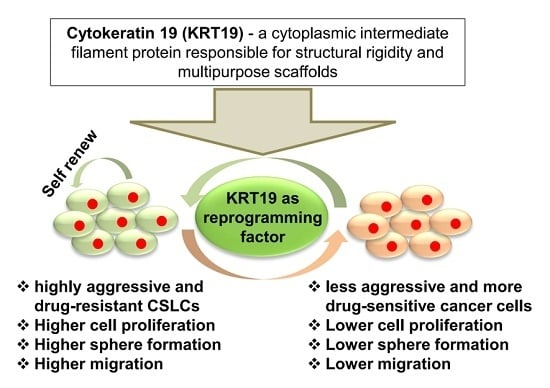
Cytokeratin 19 (KRT19) has a Role in the Reprogramming of Cancer Stem Cell-Like Cells to Less Aggressive and More Drug-Sensitive Cells
Abstract
:1. Introduction
2. Results
2.1. Expression of Cytokeratin 19 (KRT19) Is Downregulated in Several Breast Cancer Patients and Correlated with Breast Cancer Prognosis
2.2. Chemo-Treated Breast Cancer Patient-Derived KU-CSLCs Show High Expression of ALDH1, CXCR4, and CD133, and More Aggressive Cancer Phenotypes
2.3. Knockdown or Overexpression of KRT19 Regulates Cell Proliferation and Sphere Formation in MDA-MB231 and KU-CSLC Cells
2.4. Silencing or Upregulation of KRT19 Regulates Cell Migration in MDA-MB231 and KU-CSLC Cells
2.5. KRT19 Can Regulate Drug Resistance Capacity in Breast Cancer Cells
2.6. KRT19 Regulates Cancer Stem Cell Reprogramming through p-GSK3β (Tyr216) and p-Src Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Human Tissue Samples
4.2. Bioinformatics Analysis
4.3. Cell Culture
4.4. Total RNA Extraction and Reverse Transcriptase-PCR (RT-PCR) Analyses
4.5. Polymerase Chain Reaction (PCR) and Quantitative Real-Time-PCR (qRT-PCR)
4.6. In Vivo Tumorigenicity Analysis
4.7. Knockdown and Overexpression of KRT19
4.8. Western Blotting (WB) Analysis
4.9. Cell Proliferation Analysis
4.10. Sphere Formation Assay
4.11. Wound-Healing/Migration Assay
4.12. Drug-Resistance Assay
4.13. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| KRT19 | cytokeratin 19 |
| iPSC | induced pluripotent stem cell |
| PS | pluripotent stem |
| CSC | cancer stem cell |
| KU-CSLC | konkuk University-cancer stem cell-like cell |
| KRT19-oe | keratin 19-overexpression |
| RT-PCR | reverse transcriptase polymerase chain reaction |
| TF | Transcription factor |
| qRT-PCR | quantitative real-time-PCR |
| shRNA | short hairpin RNA |
| shKRT19 | short hairpin keratin19 |
| SCID | severe combined immunodeficiency |
| WB | western blot |
| EMT | epithelial to mesenchymal transition |
| MET | mesenchymal to epithelial transition |
| DOX | doxorubicin |
| STR | short tandem repeat |
| CDS | coding sequence |
| SDS-PAGE | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| TBS | tris-buffered saline |
| PBS | phosphate-buffered saline |
| TBST | tris-buffered saline Tween 20 |
| HRP | horseradish peroxidase |
| ECL | enhanced chemiluminescence |
References
- Almendro, V.; Marusyk, A.; Polyak, K. Cellular Heterogeneity and Molecular Evolution in Cancer. Annu. Rev. Pathol. Mech. 2013, 8, 277–302. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- You, J.S.; Jones, P.A. Cancer genetics and epigenetics: Two sides of the same coin? Cancer Cell 2012, 22, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Chestkov, I.V.; Khomyakova, E.A.; Vasilieva, E.A.; Lagarkova, M.A.; Kiselev, S.L. Molecular barriers to processes of genetic reprogramming and cell transformation. Biochemistry 2014, 79, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- Bogomazova, A.N.; Vassina, E.M.; Kiselev, S.I.; Lagarkova, M.A.; Lebedeva, O.S.; Nekrasov, E.D.; Panova, A.V.; Philonenko, E.S.; Khomyakova, E.A.; Tskhovrebova, L.V.; et al. Genetic Cell Reprogramming: A New Technology for Basic Research and Applied Usage. Genetika 2015, 51, 466–478. [Google Scholar] [CrossRef] [PubMed]
- Hochedlinger, K.; Yamada, Y.; Beard, C.; Jaenisch, R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 2005, 121, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Chiou, S.H.; Wang, M.L.; Chou, Y.T.; Chen, C.J.; Hong, C.F.; Hsieh, W.J.; Chang, H.T.; Chen, Y.S.; Lin, T.W.; Hsu, H.S.; et al. Coexpression of Oct4 and Nanog Enhances Malignancy in Lung Adenocarcinoma by Inducing Cancer Stem Cell-Like Properties and Epithelial-Mesenchymal Transdifferentiation. Cancer Res. 2010, 70, 10433–10444. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monk, M.; Holding, C. Human embryonic genes re-expressed in cancer cells. Oncogene 2001, 20, 8085–8091. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Ji, X.; Zhang, F.; Li, L.; Ma, L. Embryonic Stem Cell Markers. Molecules 2012, 17, 6196. [Google Scholar] [CrossRef] [PubMed]
- Schoenhals, M.; Kassambara, A.; De Vos, J.; Hose, D.; Moreaux, J.; Klein, B. Embryonic stem cell markers expression in cancers. Biochem. Biophs. Res. Commun. 2009, 383, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Gillis, A.J.M.; Stoop, H.; Biermann, K.; van Gurp, R.J.; Swartzman, E.; Cribbes, S.; Ferlinz, A.; Shannon, M.; Oosterhuis, J.W.; Looijenga, L.H.J. Expression and interdependencies of pluripotency factors LIN28, OCT3/4, NANOG and SOX2 in human testicular germ cells and tumours of the testis. Int. J. Androl. 2011, 34, E160–E174. [Google Scholar] [CrossRef] [PubMed]
- Forghanifard, M.M.; Khales, S.A.; Javdani-Mallak, A.; Rad, A.; Farshchian, M.; Abbaszadegan, M.R. Stemness state regulators SALL4 and SOX2 are involved in progression and invasiveness of esophageal squamous cell carcinoma. Med. Oncol. 2014, 31, 922. [Google Scholar] [CrossRef] [PubMed]
- Sodja, E.; Rijavec, M.; Koren, A.; Sadikov, A.; Korosec, P.; Cufer, T. The prognostic value of whole blood SOX2, NANOG and OCT4 mRNA expression in advanced small-cell lung cancer. Radiol. Oncol. 2016, 50, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Tai, S.K.; Yang, M.H.; Chang, S.Y.; Chang, Y.C.; Li, W.Y.; Tsai, T.L.; Wang, Y.F.; Chu, P.Y.; Hsieh, S.L. Persistent Kruppel-like factor 4 expression predicts progression and poor prognosis of head and neck squamous cell carcinoma. Cancer Sci. 2011, 102, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Loven, J.; Rahl, P.B.; Paranal, R.M.; Burge, C.B.; Bradner, J.E.; Lee, T.I.; Young, R.A. Transcriptional Amplification in Tumor Cells with Elevated c-Myc. Cell 2012, 151, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Zhu, Z.; Zeng, F. Expression and significance of Oct4 in bladder cancer. J. Huazhong Univ. Sci. Technol. Med. Sci. 2007, 27, 675–677. [Google Scholar] [CrossRef] [PubMed]
- Hatefi, N.; Nouraee, N.; Parvin, M.; Ziaee, S.A.M.; Mowla, S.J. Evaluating the Expression of Oct4 as a Prognostic Tumor Marker in Bladder Cancer. Iran. J. Basic Med. Sci. 2012, 15, 1154–1161. [Google Scholar] [PubMed]
- De Resende, M.F.; Chinen, L.T.D.; Vieira, S.; Jampietro, J.; da Fonseca, F.P.; Vassallo, J.; Campos, L.C.; Guimares, G.C.; Soares, F.A.; Rocha, R.M. Prognostication of OCT4 isoform expression in prostate cancer. Tumor Biol. 2013, 34, 2665–2673. [Google Scholar] [CrossRef] [PubMed]
- Rodini, C.O.; Suzuki, D.E.; Saba-Silva, N.; Cappellano, A.; de Souza, J.E.S.; Cavalheiro, S.; Toledo, S.R.C.; Okamoto, O.K. Expression analysis of stem cell-related genes reveal OCT4 as a predictor of poor clinical outcome in medulloblastoma. J. Neuro-Oncol. 2012, 106, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Li, C.G.; Yan, Y.; Ji, W.D.; Bao, L.L.; Qian, H.H.; Chen, L.; Wu, M.C.; Chen, H.Z.; Li, Z.G.; Su, C.Q. OCT4 Positively Regulates Survivin Expression to Promote Cancer Cell Proliferation and Leads to Poor Prognosis in Esophageal Squamous Cell Carcinoma. PLoS ONE 2012, 7, e49693. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, K.; Semi, K.; Yamamoto, T.; Shimizu, M.; Tanaka, A.; Mitsunaga, K.; Okita, K.; Osafune, K.; Arioka, Y.; Maeda, T.; et al. Premature Termination of Reprogramming In Vivo Leads to Cancer Development through Altered Epigenetic Regulation. Cell 2014, 156, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar] [PubMed]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.T.; Berry, P.A.; Hyde, C.; Stower, M.J.; Maitland, N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005, 65, 10946–10951. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.F.; Dick, J.E.; Dirks, P.B.; Eaves, C.J.; Jamieson, C.H.; Jones, D.L.; Visvader, J.; Weissman, I.L.; Wahl, G.M. Cancer stem cells––Perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006, 66, 9339–9344. [Google Scholar] [CrossRef] [PubMed]
- Pardal, R.; Clarke, M.F.; Morrison, S.J. Applying the principles of stem-cell biology to cancer. Nat. Rev. Cancer 2003, 3, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Xenidis, N.; Perraki, M.; Apostolaki, S.; Politaki, E.; Kafousi, M.; Stathopoulos, E.N.; Stathopoulou, A.; Lianidou, E.; Chlouverakis, G. Different prognostic value of cytokeratin-19 mRNA positive circulating tumor cells according to estrogen receptor and HER2 status in early-stage breast cancer. J. Clin. Oncol. 2007, 25, 5194–5202. [Google Scholar] [CrossRef] [PubMed]
- Bozionellou, V.; Mavroudis, D.; Perraki, M.; Papadopoulos, S.; Apostolaki, S.; Stathopoulos, E.; Stathopoulou, A.; Lianidou, E.; Georgoulias, V. Trastuzumab administration can effectively target chemotherapy-resistant cytokeratin-19 messenger RNA–positive tumor cells in the peripheral blood and bone marrow of patients with breast cancer. Clin. Cancer Res. 2004, 10, 8185–8194. [Google Scholar] [CrossRef] [PubMed]
- Fradette, J.; Germain, L.; Seshaiah, P.; Coulombe, P.A. The type I keratin 19 possesses distinct and context-dependent assembly properties. J. Biol. Chem. 1998, 273, 35176–35184. [Google Scholar] [CrossRef] [PubMed]
- Coulombe, P.A.; Wong, P. Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat. Cell Biol. 2004, 6, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, M.J.C.; Seftor, E.A.; Chu, Y.W.; Trevor, K.T.; Seftor, R.E.B. Role of intermediate filaments in migration, invasion and metastasis. Cancer Metast. Rev. 1996, 15, 507–525. [Google Scholar] [CrossRef]
- Kawai, T.; Yasuchika, K.; Ishii, T.; Katayama, H.; Yoshitoshi, E.Y.; Ogiso, S.; Kita, S.; Yasuda, K.; Fukumitsu, K.; Mizumoto, M.; et al. Keratin 19, a Cancer Stem Cell Marker in Human Hepatocellular Carcinoma. Clin. Cancer Res. 2015, 21, 3081–3091. [Google Scholar] [CrossRef] [PubMed]
- Govaere, O.; Petz, M.; Wouters, J.; Vandewynckel, Y.-P.; Scott, E.J.; Topal, B.; Nevens, F.; Verslype, C.; Anstee, Q.M.; Van Vlierberghe, H. The PDGFRα-laminin B1-keratin 19 cascade drives tumor progression at the invasive front of human hepatocellular carcinoma. Oncogene 2017, 36, 6605–6616. [Google Scholar] [CrossRef] [PubMed]
- Govaere, O.; Komuta, M.; Berkers, J.; Spee, B.; Janssen, C.; de Luca, F.; Katoonizadeh, A.; Wouters, J.; van Kempen, L.C.; Durnez, A.; et al. Keratin 19: A key role player in the invasion of human hepatocellular carcinomas. Gut 2014, 63, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Choi, G.H.; Na, D.C.; Ahn, E.Y.; Kim, G.I.; Lee, J.E.; Cho, J.Y.; Yoo, J.E.; Choi, J.S.; Park, Y.N. Human hepatocellular carcinomas with “Stemness”-related marker expression: Keratin 19 expression and a poor prognosis. Hepatology 2011, 54, 1707–1717. [Google Scholar] [CrossRef] [PubMed]
- Ju, J.H.; Oh, S.; Lee, K.M.; Yang, W.; Nam, K.S.; Moon, H.G.; Noh, D.Y.; Kim, C.G.; Park, G.; Park, J.B.; et al. Cytokeratin19 induced by HER2/ERK binds and stabilizes HER2 on cell membranes. Cell Death Differ. 2015, 22, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, T.; Sakaguchi, M.; Yamamoto, H.; Tomida, S.; Takata, K.; Shien, K.; Hashida, S.; Miyata-Takata, T.; Watanabe, M.; Suzawa, K.; et al. Interaction of cytokeratin 19 head domain and HER2 in the cytoplasm leads to activation of HER2-Erk pathway. Sci. Rep. 2016, 6, 39557. [Google Scholar] [CrossRef] [PubMed]
- Lapouge, G.; Youssef, K.K.; Vokaer, B.; Achouri, Y.; Michaux, C.; Sotiropoulou, P.A.; Blanpain, C. Identifying the cellular origin of squamous skin tumors. Proc. Natl. Acad. Sci. USA 2011, 108, 7431–7436. [Google Scholar] [CrossRef] [PubMed]
- Means, A.L.; Xu, Y.W.; Zhao, A.Z.; Ray, K.C.; Gu, G.Q. A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis 2008, 46, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Asfaha, S.; Hayakawa, Y.; Muley, A.; Stokes, S.; Graham, T.A.; Ericksen, R.E.; Westphalen, C.B.; von Burstin, J.; Mastracci, T.L.; Worthley, D.L.; et al. Krt19(+)/Lgr5(-) Cells Are Radioresistant Cancer-Initiating Stem Cells in the Colon and Intestine. Cell Stem Cell 2015, 16, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Kabir, N.N.; Ronnstrand, L.; Kazi, J.U. Keratin 19 expression correlates with poor prognosis in breast cancer. Mol. Biol. Rep. 2014, 41, 7729–7735. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Choi, H.; Kim, B.; Dayem, A.; Yang, G.; Kim, K.; Yin, Y.; Cho, S. KRT19 directly interacts with β-catenin/RAC1 complex to regulate NUMB-dependent NOTCH signaling pathway and breast cancer properties. Oncogene 2017, 36, 332–349. [Google Scholar] [CrossRef] [PubMed]
- Ju, J.H.; Yang, W.; Lee, K.M.; Oh, S.; Nam, K.; Shim, S.; Shin, S.Y.; Gye, M.C.; Chu, I.S.; Shin, I. Regulation of Cell Proliferation and Migration by Keratin19-Induced Nuclear Import of Early Growth Response-1 in Breast Cancer Cells. Clin. Cancer Res. 2013, 19, 4335–4346. [Google Scholar] [CrossRef] [PubMed]
- Bambang, I.F.; Lu, D.; Li, H.P.; Chiu, L.L.; Lau, Q.C.; Koay, E.; Zhang, D.H. Cytokeratin 19 regulates endoplasmic reticulum stress and inhibits ERp29 expression via p38 MAPK/XBP-1 signaling in breast cancer cells. Exp. Cell Res. 2009, 315, 1964–1974. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Miki, J.; Furusato, B.; Li, H.; Gu, Y.; Takahashi, H.; Egawa, S.; Sesterhenn, I.A.; McLeod, D.G.; Srivastava, S.; Rhim, J.S. Identification of putative stem cell markers, CD133 and CXCR4, in hTERT-immortalized primary nonmalignant and malignant tumor-derived human prostate epithelial cell lines and in prostate cancer specimens. Cancer Res. 2007, 67, 3153–3161. [Google Scholar] [CrossRef] [PubMed]
- Douville, J.; Beaulieu, R.; Balicki, D. ALDH1 as a Functional Marker of Cancer Stem and Progenitor Cells. Stem Cells Dev. 2009, 18, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Martini, M.; De Santis, M.C.; Braccini, L.; Gulluni, F.; Hirsch, E. PI3K/AKT signaling pathway and cancer: An updated review. Ann. Med. 2014, 46, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiang, G.; Sun, M.; Zhou, H.; Miao, Y.; Liang, M.; Wang, E.; Zhang, Y. Cytosolic THUMPD1 promotes breast cancer cells invasion and metastasis via the AKT-GSK3-Snail pathway. Oncotarget 2017, 8, 13357–13366. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, W.Z.; Liu, T.; Feng, X.; Yang, N.; Zhou, H.F. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal. Transduct. Res. 2015, 35, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.K.; Cui, Y.H.; Yoo, K.C.; Kim, I.G.; Lee, M.; Choi, Y.H.; Suh, Y.; Lee, S.J. Radiation promotes malignant phenotypes through SRC in breast cancer cells. Cancer Sci. 2015, 106, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Malki, A.; Elbayaa, R.Y.; Ashour, H.M.A.; Loffredo, C.A.; Youssef, A.M. Novel thiosemicarbazides induced apoptosis in human MCF-7 breast cancer cells via JNK signaling. J. Enzym. Inhib. Med. Chem. 2015, 30, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Li, J.; Chen, H.; Fu, J.; Ray, S.; Huang, S.; Zheng, H.; Ai, W. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene 2011, 30, 2161–2172. [Google Scholar] [CrossRef] [PubMed]
- Leis, O.; Eguiara, A.; Lopez-Arribillaga, E.; Alberdi, M.J.; Hernandez-Garcia, S.; Elorriaga, K.; Pandiella, A.; Rezola, R.; Martin, A.G. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 2012, 31, 1354–1365. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.R.; Yu, J.J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Varambally, R.; Yu, J.J.; Briggs, B.B.; Barrette, T.R.; Anstet, M.J.; Kincead-Beal, C.; Kulkarni, P.; et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007, 9, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Mansour, S.F.; Atwa, M.M. Clinicopathological Significance of CD133 and ALDH1 Cancer Stem Cell Marker Expression in Invasive Ductal Breast Carcinoma. Asian Pac. J. Cancer Prev. 2015, 16, 7491–7496. [Google Scholar] [CrossRef] [PubMed]
- Roudi, R.; Korourian, A.; Shariftabrizi, A.; Madjd, Z. Differential Expression of Cancer Stem Cell Markers ALDH1 and CD133 in Various Lung Cancer Subtypes. Cancer Invest. 2015, 33, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.; D’Alterio, C.; Camerlingo, R.; Tirino, V.; Consales, C.; Riccio, A.; Ierano, C.; Cecere, S.C.; Losito, N.S.; Greggi, S.; et al. Identification of a distinct population of CD133+CXCR4+ cancer stem cells in ovarian cancer. Sci. Rep. 2015, 5, 10357. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.B.; Xie, S.P.; Xiong, M.; Liu, Y.C.; Yang, X.Y.; Tembo, K.M.; Huang, J.; Hu, W.D.; Huang, X.X.; Pan, S.; et al. CXCR4 is involved in CD133-induced EMT in non-small cell lung cancer. Int. J. Oncol. 2017, 50, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yoshida, T.; Okabe, M.; Zhou, K.X.; Wang, F.; Soko, C.; Saito, S.; Nikaido, T. Isolation of Stem-Like Cancer Cells in Primary Endometrial Cancer Using Cell Surface Markers CD133 and CXCR4. Transl. Oncol. 2017, 10, 976–987. [Google Scholar] [CrossRef] [PubMed]
- Adorno-Cruz, V.; Kibria, G.; Liu, X.; Doherty, M.; Junk, D.J.; Guan, D.; Hubert, C.; Venere, M.; Mulkearns-Hubert, E.; Sinyuk, M.; et al. Cancer stem cells: Targeting the roots of cancer, seeds of metastasis, and sources of therapy resistance. Cancer Res. 2015, 75, 924–929. [Google Scholar] [CrossRef] [PubMed]
- Li, C.G.; Zhu, M.L.; Lou, X.L.; Liu, C.Y.; Chen, H.Z.; Lin, X.J.; Ji, W.D.; Li, Z.G.; Su, C.Q. Transcriptional factor OCT4 promotes esophageal cancer metastasis by inducing epithelial-mesenchymal transition through VEGF-C/VEGFR-3 signaling pathway. Oncotarget 2017, 8, 71933–71945. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.C.; Xie, F.; Gao, A.D.; Zhang, R.; Zhang, L.; Xiao, Z.W.; Hu, Q.; Huang, W.F.; Huang, Q.J.; Lin, B.S.; et al. SOX2 regulates multiple malignant processes of breast cancer development through the SOX2/miR-181a-5p, miR-30e-5p/TUSC3 axis. Mol. Cancer 2017, 16, 62. [Google Scholar] [CrossRef] [PubMed]
- Murgai, M.; Ju, W.; Eason, M.; Kline, J.; Kaplan, R.N. KLF4-dependent perivascular plasticity enhances pre-metastatic niche formation and metastasis. Nat. Med. 2017, 23, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Kong, J.; Liu, Y.; Li, Z.; Xia, J.; Zhang, Y.; Zhao, S.; Li, F.; Li, J.; Gu, C. Transcriptional activation of NANOG by YBX1 promotes lung cancer stem-like properties and metastasis. Biochem. Biophys. Res. Commun. 2017, 487, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Hsu, H.-S.; Chen, Y.-W.; Tsai, T.-H.; How, C.-K.; Wang, C.-Y.; Hung, S.-C.; Chang, Y.-L.; Tsai, M.-L.; Lee, Y.-Y. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS ONE 2008, 3, e2637. [Google Scholar] [CrossRef] [PubMed]
- Saigusa, S.; Tanaka, K.; Toiyama, Y.; Yokoe, T.; Okugawa, Y.; Ioue, Y.; Miki, C.; Kusunoki, M. Correlation of CD133, OCT4, and SOX2 in rectal cancer and their association with distant recurrence after chemoradiotherapy. Ann. Surg. Oncol. 2009, 16, 3488–3498. [Google Scholar] [CrossRef] [PubMed]
- Mu, P.; Zhang, Z.; Benelli, M.; Karthaus, W.R.; Hoover, E.; Chen, C.-C.; Wongvipat, J.; Ku, S.-Y.; Gao, D.; Cao, Z. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53-and RB1-deficient prostate cancer. Science 2017, 355, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Riz, I.; Hawley, T.S.; Hawley, R.G. KLF4-SQSTM1/p62-associated prosurvival autophagy contributes to carfilzomib resistance in multiple myeloma models. Oncotarget 2015, 6, 14814–14831. [Google Scholar] [CrossRef] [PubMed]
- Tsai, L.L.; Yu, C.C.; Chang, Y.C.; Yu, C.H.; Chou, M.Y. Markedly increased Oct4 and Nanog expression correlates with cisplatin resistance in oral squamous cell carcinoma. J. Oral Pathol. Med. 2011, 40, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Bueno, G.; Portillo, F.; Cano, A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene 2008, 27, 6958–6969. [Google Scholar] [CrossRef] [PubMed]
- De Figueiredo-Pontes, L.L.; Pintao, M.C.T.; Oliveira, L.C.O.; Dalmazzo, L.F.F.; Jacomo, R.H.; Garcia, A.B.; Falcao, R.P.; Rego, E.M. Determination of P-glycoprotein, MDR-related protein 1, breast cancer resistance protein, and lung-resistance protein expression in leukemic stem cells of acute myeloid leukemia. Cytom. B-Clin. Cytom. 2008, 74B, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Goding, C.R.; Pei, D.; Lu, X. Cancer: Pathological nuclear reprogramming? Nat. Rev. Cancer 2014, 14, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Nuclear reprogramming and the cancer genome. Nat. Genet. 2013, 45, 963.
- Kurebayashi, J.; Kanomata, N.; Moriya, T.; Kozuka, Y.; Watanabe, M.; Sonoo, H. Preferential antitumor effect of the Src inhibitor dasatinib associated with a decreased proportion of aldehyde dehydrogenase 1-positive cells in breast cancer cells of the basal B subtype. BMC Cancer 2010, 10, 568. [Google Scholar] [CrossRef] [PubMed]
- Bolos, V.; Gasent, J.M.; Lopez-Tarruella, S.; Grande, E. The dual kinase complex FAK-Src as a promising therapeutic target in cancer. OncoTargets Ther. 2010, 3, 83–97. [Google Scholar] [CrossRef]
- Orgaz, J.L.; Pandya, P.; Dalmeida, R.; Karagiannis, P.; Sanchez-Laorden, B.; Viros, A.; Albrengues, J.; Nestle, F.O.; Ridley, A.J.; Gaggioli, C.; et al. Diverse matrix metalloproteinase functions regulate cancer amoeboid migration. Nat. Commun. 2014, 5, 4255. [Google Scholar] [CrossRef] [PubMed]
- Thakur, R.; Trivedi, R.; Rastogi, N.; Singh, M.; Mishra, D.P. Inhibition of STAT3, FAK and Src mediated signaling reduces cancer stem cell load, tumorigenic potential and metastasis in breast cancer. Sci. Rep. 2015, 5, 10194. [Google Scholar] [CrossRef] [PubMed]
- Paladino, D.; Yue, P.; Furuya, H.; Acoba, J.; Rosser, C.J.; Turkson, J. A novel nuclear Src and p300 signaling axis controls migratory and invasive behavior in pancreatic cancer. Oncotarget 2016, 7, 7253–7267. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Fang, J.; Ding, X.J.; Chen, Q.Y. Role and inhibition of Src signaling in the progression of liver cancer. Open Life Sci. 2016, 11, 513–518. [Google Scholar] [CrossRef]
- Goc, A.; Al-Husein, B.; Katsanevas, K.; Steinbach, A.; Lou, U.; Sabbineni, H.; DeRemer, D.L.; Somanath, P.R. Targeting Src-mediated Tyr216 phosphorylation and activation of GSK-3 in prostate cancer cells inhibit prostate cancer progression in vitro and in vivo. Oncotarget 2014, 5, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Jope, R.S.; Yuskaitis, C.J.; Beurel, E. Glycogen synthase kinase-3 (GSK3): Inflammation, diseases, and therapeutics. Neurochem. Res. 2007, 32, 577–595. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.; Nikolakaki, E.; Plyte, S.E.; Totty, N.F.; Woodgett, J.R. Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. EMBO J. 1993, 12, 803–808. [Google Scholar] [PubMed]
- Kaidanovich-Beilin, O.; Woodgett, J.R. GSK-3: Functional Insights from Cell Biology and Animal Models. Front. Mol. Neurosci. 2011, 4, 40. [Google Scholar] [CrossRef] [PubMed]
- Lánczky, A.; Nagy, Á.; Bottai, G.; Munkácsy, G.; Szabó, A.; Santarpia, L.; Győrffy, B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 2016, 160, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Dayem, A.A.; Kim, B.; Gurunathan, S.; Choi, H.Y.; Yang, G.; Saha, S.K.; Han, D.; Han, J.; Kim, K.; Kim, J.H.; et al. Biologically synthesized silver nanoparticles induce neuronal differentiation of SH-SY5Y cells via modulation of reactive oxygen species, phosphatases, and kinase signaling pathways. Biotechnol. J. 2014, 9, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, S.; Konno, M.; Hamabe, A.; Hasegawa, S.; Kano, Y.; Fukusumi, T.; Satoh, T.; Takiguchi, S.; Mori, M.; Doki, Y.; et al. Surgically resected human tumors reveal the biological significance of the gastric cancer stem cell markers CD44 and CD26. Oncol. Lett. 2015, 9, 2361–2367. [Google Scholar] [CrossRef] [PubMed]
- Feldman, J.P.; Goldwasser, R.; Mark, S.; Schwartz, J.; Orion, I. A mathematical model for tumor volume evaluation using two-dimensions. J. Appl. Quant. Methods 2009, 4, 455–462. [Google Scholar]
- Kutner, R.H.; Zhang, X.Y.; Reiser, J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 2009, 4, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.K.; Yin, Y.; Kim, K.; Yang, G.M.; Dayem, A.A.; Choi, H.Y.; Cho, S.G. Valproic Acid Induces Endocytosis-Mediated Doxorubicin Internalization and Shows Synergistic Cytotoxic Effects in Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2017, 18, 1048. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.H.; Kim, D.K.; Cha, Y.; Jeon, I.; Song, J.; Park, K.S. PI3K/Akt and Stat3 signaling regulated by PTEN control of the cancer stem cell population, proliferation and senescence in a glioblastoma cell line. Int. J. Oncol. 2013, 42, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Kujawski, M.; Kortylewski, M.; Lee, H.; Herrmann, A.; Kay, H.; Yu, H. Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J. Clin. Investig. 2008, 118, 3367–3377. [Google Scholar] [CrossRef] [PubMed]
- Hua, G.J.; Liu, Y.P.; Li, X.Y.; Xu, P.R.; Luo, Y.C. Targeting glucose metabolism in chondrosarcoma cells enhances the sensitivity to doxorubicin through the inhibition of lactate dehydrogenase-A. Oncol. Rep. 2014, 31, 2727–2734. [Google Scholar] [CrossRef] [PubMed]







| Accession No. | Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|---|
| NM_000689.4 | ALDH1 | CTGCTGGCGACAATGGAGT | GTCAGCCCAACCTGCACAG |
| NM_001008540.2 | CXCR4 | CGCCACCAACAGTCAGAG | AACACAACCACCCACAAGTC |
| NM_001145852.1 | CD133 | GTCACCATTGACTTCTTGGTGCTGT | TGTCAGATGGAGTTACGCAGGTTTC |
| NM_002046.5 | GAPDH | AATCCCATCACCATCTTCCAG | CACGATACCAAAGTTGTCATGG |
| NM_002276.4 | KRT19 | GCGAGCTAGAGGTGAAGATC | CGGAAGTCATCTGCAGCCA |
| NM_004827.2 | ABCG2 | TTATCCGTGGTGTGTCTGGAG | TCCTGCTTGGAAGGCTCTATG |
| NM_004996.3 | ABCC1 | GCCGGTGAAGGTTGTGTACT | CTGACGAAGCAGATGTGGAA |
| NM_001348946.1 | ABCB1 | GAGGAAGACATGACCAGGTA | CTGTCGCATTATAGCATGAA |
| NM_001285986.1 | OCT4 | GTCCCAGGACATCAAAGCTC | CTCCAGGTTGCCTCTCACTC |
| NM_002701.5 | OCT4A | CGTGAAGCTGGAGAAGGAGAAGCTG | CAAGGGCCGCAGCTTACACATGTTC |
| NM_001285987.1 | OCT4B | ATGCATGAGTCAGTGAACAG | CCACATCGGCCTGTGTATAT |
| NM_001314052.1 | KLF4 | GAACTGACCAGGCACTACCG | TTCTGGCAGTGTGGGTCATA |
| NM_003106.3 | SOX2 | ACACCAATCCCATCCACACT | GCAAGAAGCCTCTCCTTGAA |
| NM_024865.3 | NANOG | ATACCTCAGCCTCCAGCAGA | GCAGGACTGCAGAGATTCCT |
| NM_002467.4 | C-MYC | CTCGGATTCTCTGCTCTC | TCGCCTCTTGACATTCTC |
| NM_001126118.1 | P53 | GCCCAACAACACCAGCTCCT | CCTGGGCATCCTTGAGTTCC |
| XM_017008921.2 | N-cadherin | TGGATGGACCTTATGTTGCT | AACACCTGTCTTGGGATCAA |
| NM_001205255.1 | OCLN | CTTCAGGCAGCCTCGTTACA | TCCTCCTCCAGCTCATCACA |
| NM_001317185.1 | E-cadherin | CAG CAC GTA CAC AGC CCT AA | ACC CAC CTC TAA GGC CAT CT |
| NM_001323654.1 | ZEB1 | GCCAATAAGCAAACGATTCTG | TTTGGCTGGATCACTTTCAAG |
| NM_000474.3 | TWIST1 | CTCAGCTACGCCTTCTCG | ACTGTCCATTTTCTCCTTCTCTG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saha, S.K.; Kim, K.; Yang, G.-M.; Choi, H.Y.; Cho, S.-G. Cytokeratin 19 (KRT19) has a Role in the Reprogramming of Cancer Stem Cell-Like Cells to Less Aggressive and More Drug-Sensitive Cells. Int. J. Mol. Sci. 2018, 19, 1423. https://doi.org/10.3390/ijms19051423
Saha SK, Kim K, Yang G-M, Choi HY, Cho S-G. Cytokeratin 19 (KRT19) has a Role in the Reprogramming of Cancer Stem Cell-Like Cells to Less Aggressive and More Drug-Sensitive Cells. International Journal of Molecular Sciences. 2018; 19(5):1423. https://doi.org/10.3390/ijms19051423
Chicago/Turabian StyleSaha, Subbroto Kumar, Kyeongseok Kim, Gwang-Mo Yang, Hye Yeon Choi, and Ssang-Goo Cho. 2018. "Cytokeratin 19 (KRT19) has a Role in the Reprogramming of Cancer Stem Cell-Like Cells to Less Aggressive and More Drug-Sensitive Cells" International Journal of Molecular Sciences 19, no. 5: 1423. https://doi.org/10.3390/ijms19051423
APA StyleSaha, S. K., Kim, K., Yang, G. -M., Choi, H. Y., & Cho, S. -G. (2018). Cytokeratin 19 (KRT19) has a Role in the Reprogramming of Cancer Stem Cell-Like Cells to Less Aggressive and More Drug-Sensitive Cells. International Journal of Molecular Sciences, 19(5), 1423. https://doi.org/10.3390/ijms19051423