Genomic Regions Analysis of Seedling Root Traits and Their Regulation in Responses to Phosphorus Deficiency Tolerance in CSSL Population of Elite Super Hybrid Rice
Abstract
:1. Introduction
2. Results
2.1. Phenotypic and Frequency Distribution of 75 CSSLs and Their Parents
2.2. Correlation Coefficient Analysis among Seedling Root Traits
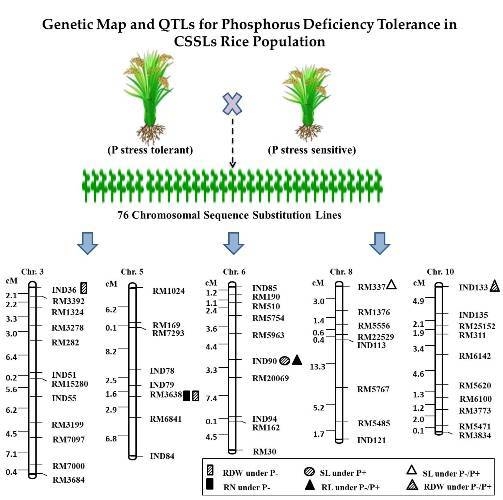
2.3. QTL Analysis for Seedling Root Traits under P+ and P− Levels and Their Relative Ratio
2.4. Seedling Root Traits in CSSLs under P deficiency
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Hydroponic Experiment
4.3. Phenotypic Investigation
4.4. QTL Mapping and Data Analysis
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Ismail, A.M.; Heuer, S.; Thomson, M.J.; Wissuwa, M. Genetic and genomic approaches to develop rice germplasm for problem soils. Plant Mol. Biol. 2007, 65, 547–570. [Google Scholar] [CrossRef] [PubMed]
- De Datta, S.K. Principles and Practice of Rice Production; John Wiley and Sons: Singapore, 1981; pp. 348–419. [Google Scholar]
- Septiningsih, E.M.; Pamplona, A.M.; Sanchez, D.L.; Neeraja, C.N.; Vergara, G.V.; Heuer, S.; Ismail, A.M.; Mackill, D.J. Development of submergence-tolerant rice cultivars: The Sub1 locus and beyond. Ann. Bot. 2009, 103, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Lea, P.J.; Miflin, B.J. Nitrogen assimilation and its relevance to crop improvement. Ann. Plant Revi. 2010, 42, 1–40. [Google Scholar]
- Cheng, L.Y.; Bucciarelli, B.; Shen, J.B.; Allan, D.; Vance, C.P. Update on white lupin cluster root acclimation to phosphorus deficiency. Plant Physiol. 2011, 156, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.L.; Wu, P.; Ling, H.Q.; Xu, G.H.; Xu, F.S.; Zhang, Q.F. Plant nutriomics in China: An overview. Ann. Bot. 2006, 98, 473–482. [Google Scholar] [CrossRef] [PubMed]
- White, P.J.; Hammond, J.P. Phosphorus nutrition of terrestrial plants. In The Ecophysiology of Plant-Phosphorus Interactions; White, P.J., Hammond, J.P., Eds.; Springer: Dordrecht, The Netherlands; pp. 51–81.
- Lambers, H.; Shane, M.W.; Cramer, M.D.; Pearse, S.J.; Veneklaas, E.J. Root structure and functioning for efficient acquisition of phosphorus: Matching morphological and physiological traits. Ann. Bot. 2006, 98, 693–713. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P. Root phenes for enhanced soil exploration and phosphorus acquisition: Tools for future crops. Plant Physiol. 2011, 156, 1041–1049. [Google Scholar] [CrossRef] [PubMed]
- Hufnagel, B. Duplicate and conquer: Multiple homologs of PHOSPHORUS-STARVATION TOLERANCE1 enhance phosphorus acquisition and sorghum performance on low-phosphorus soils. Plant Physiol. 2014, 166, 659–677. [Google Scholar] [CrossRef] [PubMed]
- Wissuwa, M.; Yano, M.; Ae, N. Mapping of QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Theor. Appl. Genet. 1998, 97, 777–783. [Google Scholar] [CrossRef]
- Chin, J.H.; Gamuyao, R.; Dalid, C.; Bustamam, M.; Prasetiyono, J.; Moeljopawiro, S.; Wissuwa, M.; Heuer, S. Developing rice with high yield under phosphorus deficiency: Pup1 sequence to application. Plant Physiol. 2011, 156, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Gupta, D.S.; Gupta, S.; Dubey, S.; Gupta, P.; Kumar, S. Quantitative trait loci from identification to exploitation for crop improvement. Plant Cell Rep. 2017, 36, 1187–1213. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wu, P.; Senadhira, D.; Huang, N. Mapping QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Theor. Appl. Genet. 1998, 97, 1361–1369. [Google Scholar] [CrossRef]
- Ming, F.; Zheng, X.; Mi, G.; He, P.; Zhu, L.; Zhang, F. Identification of quantitative trait loci affecting tolerance to low phosphorus in rice (Oryza Sativa L.). Chin. Sci. Bull. 2000, 45, 520–525. [Google Scholar] [CrossRef]
- Hu, B.; Wu, P.; Liao, C.; Zhang, W.; Ni, J. QTLs and epistasis underlying activity of acid phosphatase under phosphorus sufficient and deficient condition in rice (Oryza sativa L.). Plant Soil 2001, 230, 99–105. [Google Scholar] [CrossRef]
- Lang, N.T.; Buu, B.C. Mapping QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Omonrice 2006, 14, 1–9. [Google Scholar]
- Li, J.Z.; Xie, Y.; Dai, A.Y.; Liu, L.F.; Li, Z.C. Root and shoot traits responses to phosphorus deficiency and QTL analysis at seedling stage using introgression lines of rice. J. Genet. Genom. 2009, 36, 173–183. [Google Scholar] [CrossRef]
- Lin, D.Z.; Zhang, Y.J.; Zhang, J.Z.; Luo, L.J.; Dong, Y.J. Mapping of QTL for seedling growth response to phosphorus deficiency in rice (Oryza sativa L.). Genom. App. Biol. 2010, 29, 10–16. [Google Scholar]
- Paterson, A.H.; Lander, E.S.; Hewitt, J.D.; Peterson, S.; Lincoln, S.E.; Tanksley, S.D. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 1988, 335, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, A.; Yanagihara, S.; Kawasaki, S.; Ikehashi, H. Phosphorus deficiency induced root elongation and its QTL in rice (Oryza sativa L.). Theor. Appl. Genet. 2004, 109, 1361–1368. [Google Scholar] [CrossRef] [PubMed]
- Su, J.Y.; Xiao, Y.M.; Li, M.; Liu, Q.Y.; Li, B.; Tong, Y.P.; Jia, J.Z.; Li, Z.S. Mapping QTLs for phosphorus-deficiency tolerance at wheat seedling stage. Plant Soil 2006, 281, 25–36. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Anis, G.B.; Wang, R.; Wu, W.; Yu, N.; Shen, X.; Zhan, X.D.; Cheng, S.H.; Cao, L.Y. Genetic dissection of QTL against phosphate deficiency in the hybrid rice ‘Xieyou9308’. Plant Growth Regul. 2017, 84, 123–133. [Google Scholar] [CrossRef]
- Chaubey, C.N.; Senadhira, D.; Gregorio, G.B. Genetic analysis of tolerance for phosphorous deficiency in rice (Oryza sativa L.). Theor. Appl. Genet. 1994, 89, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.H.; Ding, P.; He, L.Y.; Xu, C.G. Genetic analysis of agricultural traits in rice related to phosphorus efficiency. Acta Genet. Sin. 2006, 33, 634–641. [Google Scholar] [CrossRef]
- Panigrahy, M.; Rao, D.N.; Sarla, N. Molecular mechanisms in response to phosphate starvation in rice. Biotechnol. Adv. 2009, 27, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Wissuwa, M.; Wegner, J.; Ae, N.; Yano, M. Substitution mapping of Pup1: A major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theor. Appl. Genet. 2002, 105, 890–897. [Google Scholar] [PubMed]
- Gamuyao, R.; Chin, J.H.; Pariasca, J.; Pesares, P.; Catausan, S.; Dalid, G.C.; Slamet, L.I.; Tecson, M.E.; Wissuwa, M.; Heuer, S. The protein kinase PstolI from traditional rice confers tolerance of phosphorus deficiency. Nature 2012, 488, 535–539. [Google Scholar] [CrossRef] [PubMed]
- De Dorlodot, S.; Forster, B.; Pagès, L.; Price, A.; Tuberosa, R.; Draye, X. Root system architecture: Opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 2007, 12, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Mu, P.; Huang, C.; Li, J.X.; Liu, L.F.; Li, Z.C. Yield trait variation and QTL mapping in a DH population of rice under phosphorus deficiency. Acta Agron. Sin. 2008, 34, 1137–1142. [Google Scholar]
- Wang, Y.; Sun, Y.J.; Chen, D.Y.; Yu, S.B. Analysis of quantitative trait loci in response to nitrogen and phosphorus deficiency in rice using chromosomal segment substitution lines. Acta Agron. Sin. 2009, 35, 580–587. [Google Scholar] [CrossRef]
- Xiang, C.; Ren, J.; Zhao, X.Q.; Ding, Z.S.; Zhang, J.; Wang, C.; Zhang, J.W.; Charles, A.J.; Zhang, Q.; Pang, Y.L.; et al. Genetic dissection of low phosphorus tolerance related traits using selected introgression lines in rice. Rice Sci. 2015, 22, 264–274. [Google Scholar]
- Kanbar, A.; Shashidhar, H.E.; Hittalmani, S. Mapping of QTL associated with root and related traits in DH population of rice (Oryza sativa L.). Indian J. Genet. 2002, 62, 287–290. [Google Scholar]
- Wang, H.M.; Xu, X.M.; Zhan, X.D.; Zhai, R.R.; Wu, W.M.; Shen, X.H.; Dai, G.X.; Cao, L.Y.; Cheng, S.H. Identification of qRL7, a major quantitative trait locus associated with rice root length in hydroponic conditions. Breed. Sci. 2013, 63, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zha, R.R.; Cao, L.Y.; Lin, Z.C.; We, X.H.; Cheng, S.H. QTLs for plant height and heading date in rice under two nitrogen levels. Acta Agron. Sin. 2011, 37, 1525–1532. [Google Scholar] [CrossRef]
- Niu, Y.F.; Chai, R.S.; Jin, G.L.; Wang, H.; Tang, C.X.; Zhang, Y.S. Responses of root architecture development to low phosphorus availability: A review. Ann. Bot. 2013, 112, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, A.; Kato, K.; Komatsu, A.; Motomura, K.; Ikehashi, H. Genetic analysis of root elongation induced by phosphorus deficiency in rice (Oryza sativa L.): Fine QTL mapping and multivariate analysis of related traits. Theor. Appl. Genet. 2008, 117, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Obara, M.; Tamura, W.; Ebitani, T.; Yano, M.; Sato, T.; Yamaya, T. Fine-mapping of qRL6.1, a major QTL for root length of rice seedlings grown under a wide range of NH4+ concentrations in hydroponic conditions. Theor. Appl. Genet. 2010, 121, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Dong, Y.J.; Zhang, J.Z.; Xiao, K.; Xu, J.L.; Terao, H. Mapping QTLs for deficiency phosphorus response to root-growth of rice seedling. Rice Genet. Newsl. 2010, 25, 36–37. [Google Scholar]
- Zhu, J.; Mickelson, S.M.; Kaeppler, S.M.; Lynch, J.P. Detection of quantitative trait loci for seminal root traits in maize (Zea mays L.) seedlings grown under differential phosphorus levels. Theor. Appl. Genet. 2006, 113, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Forno, D.A.; Cock, J.H.; Gomez, K.A. Laboratory Manual for Physiological Studies of Rice, 3rd ed.; IRRI: Manila, Philippines, 1976. [Google Scholar]





| Treat. | Trait | Parents | p-Value | Chromosomal Segment Substitution Lines (CSSLs) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| XieqingzaoB | Zhonghui9308 | Min. | Max. | Mean | SD% | Kurtosis | Skewness | |||
| Normal phosphorus (P+) | SL | 20.68 | 21.67 | 0.172 | 16.52 | 27.82 | 21.06 | 2.02 | 0.79 | 0.12 |
| RL | 8.93 | 7.70 | 0.071 | 4.43 | 9.23 | 6.56 | 1.14 | −0.58 | 0.20 | |
| RN | 9.00 | 7.17 | 0.022 | 5.00 | 8.67 | 6.18 | 0.80 | −0.19 | 0.47 | |
| SDW | 86.43 | 83.83 | 0.007 | 55.38 | 104.8 | 80.91 | 10.64 | −0.25 | 0.27 | |
| RDW | 37.22 | 28.00 | 3.710 | 18.33 | 39.0 | 26.59 | 5.13 | −0.46 | 0.64 | |
| TDW | 123.65 | 111.83 | 0.000 | 82.01 | 142.1 | 107.51 | 14.65 | −0.47 | 0.48 | |
| Low phosphorus (P-) | SL | 19.46 | 20.29 | 0.166 | 15.03 | 24.33 | 20.48 | 2.19 | −0.25 | −0.52 |
| RL | 7.54 | 6.34 | 0.071 | 4.37 | 10.33 | 7.62 | 1.08 | 0.39 | 0.07 | |
| RN | 7.35 | 6.00 | 0.051 | 5.67 | 9.50 | 7.36 | 0.79 | −0.37 | 0.33 | |
| SDW | 84.33 | 79.00 | 0.001 | 50.00 | 115.67 | 87.39 | 16.81 | −1.10 | −0.08 | |
| RDW | 32.67 | 26.67 | 0.029 | 19.38 | 59.00 | 33.79 | 9.84 | −0.59 | 0.46 | |
| TDW | 117.00 | 105.67 | 2.36 | 82.67 | 167.7 | 121.19 | 25.29 | −1.35 | 0.14 | |
| S.O.V | df | MS | F-Value | p-Value |
|---|---|---|---|---|
| Shoot length (cm) | ||||
| Genotype (G) | 74 | 14.92 | 17.01 | 0.01 |
| Treatment (T) | 1 | 25.61 | 29.31 | 0.34 |
| G × T | 74 | 2.93 | 33.00 | 0.73 |
| Error | 150 | 4.09 | ||
| Root length (cm) | ||||
| Genotype (G) | 74 | 2.26 | 13.06 | 0.02 |
| Treatment (T) | 1 | 84.44 | 49.00 | 0.20 |
| G × T | 74 | 2.72 | 16.60 | 0.48 |
| Error | 150 | 1.18 | ||
| Root number | ||||
| Genotype (G) | 74 | 0.70 | 19.35 | 0.11 |
| Treatment (T) | 1 | 102.80 | 28.25 | 0.00 |
| G × T | 74 | 1.85 | 50.87 | 0.08 |
| Error | 150 | 0.65 | ||
| Shoot dry weight (mg) | ||||
| Genotype (G) | 74 | 373.95 | 40.15 | 0.76 |
| Treatment (T) | 1 | 3152.26 | 33.84 | 0.00 |
| G × T | 74 | 417.90 | 44.87 | 0.27 |
| Error | 150 | 113.21 | ||
| Root dry weight (mg) | ||||
| Genotype (G) | 74 | 76.92 | 498.22 | 0.00 |
| Treatment (T) | 1 | 3889.58 | 100.23 | 0.00 |
| G × T | 74 | 169.65 | 437.00 | 0.00 |
| Error | 150 | 96.90 | ||
| Total dry weight (mg) | ||||
| Genotype (G) | 74 | 691.25 | 57.09 | 0.44 |
| Treatment (T) | 1 | 14045.53 | 113.60 | 0.90 |
| G × T | 74 | 1017.78 | 84.06 | 0.63 |
| Error | 150 | 214.63 | ||
| Low Phosphorus (P−) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Normal Phosphorus (P+) | Traits | SL | RL | RN | SDW | RDW | TDW | Traits | Low Phosphorus (P-) |
| SL | 0.673 ** | 0.519 ** | 0.394 ** | 0.542 ** | 0.242 * | 0.454 ** | SL | ||
| RL | 0.322 ** | −0.091 | 0.440 ** | 0.621 ** | 0.592 ** | 0.643 ** | RL | ||
| RN | 0.398 ** | 0.441 ** | −0.449 ** | 0.622 ** | 0.592 ** | 0.644 ** | RN | ||
| SDW | 0.605 ** | 0.610 ** | 0.577 ** | −0.061 | 0.786 ** | 0.971 ** | SDW | ||
| RDW | 0.362 ** | 0.646 ** | 0.617 ** | 0.686 ** | −0.458 ** | 0.912 ** | RDW | ||
| TDW | 0.566 ** | 0.670 ** | 0.635 ** | 0.967 ** | 0.849 ** | −0.220 | TDW | ||
| Traits | SL | RL | RN | SDW | RDW | TDW | Traits | ||
| Normal Phosphorus (P+) | |||||||||
| Trait | Chr. | Marker | QTL | Treatment | LOD Value | Additive Effect | PVE% | M (QQ) | M (qq) |
|---|---|---|---|---|---|---|---|---|---|
| SL | 6 | InD90 | qSL6 | P+ | 3.02 | 2.57 | 16.94 | 26.10 | 20.92 |
| SL | 8 | RM337 | qSL8 | P−/P+ | 3.32 | −0.11 | 18.46 | 0.76 | 0.97 |
| RL | 6 | InD90 | qRL6 | P−/P+ | 2.31 | 0.68 | 16.07 | 2.74 | 1.04 |
| RN | 5 | RM3638 | qRN5 | P− | 2.21 | 0.62 | 12.71 | 8.54 | 7.29 |
| RDW | 3 | InD36 | qRDW3 | P− | 2.00 | −6.13 | 10.82 | 22.51 | 34.77 |
| RDW | 5 | RM3638 | qRDW5 | P− | 2.37 | 8.01 | 12.66 | 48.95 | 32.94 |
| RDW | 10 | InD133 | qRDW10 | P−/P+ | 2.18 | 0.81 | 12.52 | 2.92 | 1.30 |
| Trait | Abbreviations | Unit | Trait Measurements |
|---|---|---|---|
| Shoot length | SL | cm | Measured with a ruler |
| Root length | RL | cm | Measured with a ruler |
| Root number | RN | number | Counted manually |
| Shoot dry weight | SDW | mg | Dried and weighted using a balance (1/1000 g) |
| Root dry weight | RDW | mg | Dried and weighted using a balance (1/1000 g) |
| Total dry weight | TDW | mg | SDW + RDW |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anis, G.B.; Zhang, Y.; Wang, H.; Li, Z.; Wu, W.; Sun, L.; Riaz, A.; Cao, L.; Cheng, S. Genomic Regions Analysis of Seedling Root Traits and Their Regulation in Responses to Phosphorus Deficiency Tolerance in CSSL Population of Elite Super Hybrid Rice. Int. J. Mol. Sci. 2018, 19, 1460. https://doi.org/10.3390/ijms19051460
Anis GB, Zhang Y, Wang H, Li Z, Wu W, Sun L, Riaz A, Cao L, Cheng S. Genomic Regions Analysis of Seedling Root Traits and Their Regulation in Responses to Phosphorus Deficiency Tolerance in CSSL Population of Elite Super Hybrid Rice. International Journal of Molecular Sciences. 2018; 19(5):1460. https://doi.org/10.3390/ijms19051460
Chicago/Turabian StyleAnis, Galal Bakr, Yingxin Zhang, Huimin Wang, Zihe Li, Weixun Wu, Lianping Sun, Aamir Riaz, Liyong Cao, and Shihua Cheng. 2018. "Genomic Regions Analysis of Seedling Root Traits and Their Regulation in Responses to Phosphorus Deficiency Tolerance in CSSL Population of Elite Super Hybrid Rice" International Journal of Molecular Sciences 19, no. 5: 1460. https://doi.org/10.3390/ijms19051460
APA StyleAnis, G. B., Zhang, Y., Wang, H., Li, Z., Wu, W., Sun, L., Riaz, A., Cao, L., & Cheng, S. (2018). Genomic Regions Analysis of Seedling Root Traits and Their Regulation in Responses to Phosphorus Deficiency Tolerance in CSSL Population of Elite Super Hybrid Rice. International Journal of Molecular Sciences, 19(5), 1460. https://doi.org/10.3390/ijms19051460