Mechanism of Salt-Induced Self-Compatibility Dissected by Comparative Proteomic Analysis in Brassica napus L.
Abstract
:1. Introduction
2. Results
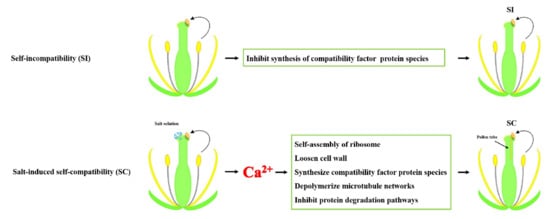
2.1. Self-Compatibility Induced by Salt Solution
2.2. Effect of Salt Solution Treatment on Self-Incompatibility-Related Gene
2.3. Quantitative Identification of Proteins Using iTRAQ
2.4. Differentially Accumulated Proteins
2.5. Functional Classification of the Differentially Accumulated Proteins
2.6. Primary Metabolic Pathways of Differentially Accumulated Proteins
2.7. Validation of Proteomics Data by Quantitative Real-Time RT-PCR
3. Discussion
3.1. Characteristics of Edible Salt Solution Destroyed SI in B. napus
3.2. Salt Stress and Salt-Induced Self-Compatibility
3.3. Protein Biosynthesis and Salt-Induced Self-Compatibility
3.4. Ubiquitin-Mediated Processes and Salt-Induced Self-Compatibility
3.5. Roles of Endocytosis and Phagosome in Salt-Induced Self-Compatibility
3.6. Plant–Pathogen Interaction and Pollen–Stigma Interaction
4. Materials and Methods
4.1. Plant Materials
4.2. Salt Solution Treatment
4.3. Aniline Blue Assay
4.4. Stigma Collection and Protein Sample Preparation
4.5. iTRAQ Labeling and SCX Fractionation
4.6. LC-MS/MS Measurement and Data Analysis
4.7. Real-Time Quantitative Reverse Transcription PCR (qPCR)
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Goring, D.R.; Banks, P.; Beversdorf, W.D.; Rothstein, S.J. Use of the polymerase chain reaction to isolate an S-locus glycoprotein cDNA introgressed from Brassica campestris into B. napus ssp. oleifera. Mol. Gen. Genet. 1992, 234, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.H. Resynthesis of Brassica napus L. for self-incompatibility: Self-incompatibility reaction, inheritance and breeding potential. Plant Breed. 2008, 124, 13–19. [Google Scholar] [CrossRef]
- Nakanishi, T.; Hinata, K. An effective time for CO2 gas treatment in overcoming self-incompatibility in Brassica. Plant Cell Physiol. 1973, 14, 873–879. [Google Scholar]
- Matsubara, S. Overcoming self-incompatibility in Raphanus sativus L. with high temperature. J. Am. Soc. Hortic. Sci. 1980, 105, 842–846. [Google Scholar]
- Okazaki, K.; Hinata, K. Repressing the expression of self-incompatibility in crucifers by short-term high temperature treatment. Theor. Appl. Genet. 1987, 73, 496–500. [Google Scholar] [CrossRef] [PubMed]
- Tatebe, T. Studies on the physiological mechanism of self-incompatibility in Japanese radish. II. J. Jpn. Soc. Horticult. Sci. 1968, 37, 43–46. [Google Scholar] [CrossRef]
- Monteiro, A.; Gabelman, W.; Williams, P. Use of sodium chloride solution to overcome self-incompatibility in Brassica campestris. HortScience 1988, 23, 876–877. [Google Scholar]
- Hu, D.; An, C.; Dong, H.; Bing, D.; Niu, J. Using chemical methods to overcome self-incompatibility of Brassica napus. Chin. Oil Crop J. 1983, 2, 23–28. [Google Scholar]
- Nettancourt, D.D. Incompatibility and Incongruity in Wild and Cultivated Plants; Springer: Berlin/Heidelberg, Germany, 2001; Volume 3, pp. 1–20. [Google Scholar]
- Bateman, A.J. Self-incompatibility systems in angiosperms. III. Cruciferae. Heredity 1955, 9, 52–68. [Google Scholar] [CrossRef]
- Takasaki, T.; Hatakeyama, K.; Suzuki, G.; Watanabe, M.; Isogai, A.; Hinata, K. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 2000, 403, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Schopfer, C.R.; Nasrallah, M.E.; Nasrallah, J.B. The Male Determinant of Self-Incompatibility in Brassica. Science 1999, 286, 1697–1700. [Google Scholar] [CrossRef] [PubMed]
- Bower, M.S.; Matias, D.D.; Fernandes-Carvalho, E.; Mazzurco, M.; Gu, T.; Rothstein, S.J.; Goring, D.R. Two members of the thioredoxin-h family interact with the kinase domain of a Brassica S locus receptor kinase. Plant Cell 1996, 8, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Haffani, Y.; Gaude, T.; Cock, J.; Goring, D. Antisense suppression of thioredoxinh mRNA in Brassica napus cv Westar pistils causes a low level constitutive pollen rejection response. Plant Mol. Biol. 2004, 55, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Kakita, M.; Murase, K.; Iwano, M.; Matsumoto, T.; Watanabe, M.; Shiba, H.; Isogai, A.; Takayama, S. Two distinct forms of M-locus protein kinase localize to the plasma membrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 2007, 19, 3961–3973. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Anderson, E.M.; Mullen, R.T.; Goring, D.R. ARC1 Is an E3 Ubiquitin Ligase and Promotes the Ubiquitination of Proteins during the Rejection of Self-Incompatible Brassica Pollen. Plant Cell 2003, 15, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.A.; Chong, Y.T.; Haasen, K.E.; Aldea-Brydges, M.G.; Stone, S.L.; Goring, D.R. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 2009, 21, 2655–2671. [Google Scholar] [CrossRef] [PubMed]
- Safavian, D.; Zayed, Y.; Indriolo, E.; Chapman, L.; Ahmed, A.; Goring, D. RNA silencing of exocyst genes in the stigma impairs the acceptance of compatible pollen in Arabidopsis. Plant Physiol. 2015, 169, 2526–2538. [Google Scholar] [CrossRef] [PubMed]
- Tantikanjana, T.; Nasrallah, M.E.; Nasrallah, J.B. Complex networks of self-incompatibility signaling in the Brassicaceae. Curr. Opin. Plant Biol. 2010, 13, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, S.; Jamshed, M.; Deb, S.; Chatfield-Reed, K.; Kwon, E.J.; Chua, G.; Samuel, M.A. Deciphering the Stigmatic Transcriptional Landscape of Compatible and Self-Incompatible Pollinations in Brassica napus Reveals a Rapid Stigma Senescence Response Following Compatible Pollination. Mol. Plant 2013, 6, 1988–1991. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Gao, C.; Yue, Y.; Liu, Z.; Ma, C.; Zhou, G.; Yang, Y.; Duan, Z.; Li, B.; Wen, J.; et al. Time-course Transcriptome Analysis of Compatible and Incompatible Pollen-Stigma Interactions in Brassica napus L. Front. Plant Sci. 2017, 8, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.A.; Tang, W.; Jamshed, M.; Northey, J.; Patel, D.; Smith, D.; Siu, K.W.; Muench, D.G.; Wang, Z.Y.; Goring, D.R. Proteomic analysis of Brassica stigmatic proteins following the self-incompatibility reaction reveals a role for microtubule dynamics during pollen responses. Mol. Cell. Proteom. 2011, 10, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, S.; Jamshed, M.; Samuel, M.A. Degradation of glyoxalase I in Brassica napus stigma leads to self-incompatibility response. Nat. Plants 2015, 1, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, S.; Jamshed, M.; Samuel, M.A. Proteomics approaches advance our understanding of Plant Self-Incompatibility response. J. Proteome Res. 2013, 12, 4717–4726. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.; Noirel, J.; Ow, S.Y.; Salim, M.; Pereira-Medrano, A.G.; Couto, N.; Pandhal, J.; Smith, D.; Pham, T.K.; Karunakaran, E.; et al. An insight into iTRAQ: Where do we stand now? Anal. Bioanal. Chem. 2012, 404, 1011–1027. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Anoop, V.; Gleddie, S.; Harris, L.J. Proteomic profiling of two maize inbreds during early gibberella ear rot infection. Proteomics 2011, 11, 3675–3684. [Google Scholar] [CrossRef] [PubMed]
- Ng, D.W.; Zhang, C.; Miller, M.; Shen, Z.; Briggs, S.P.; Chen, Z.J. Proteomic divergence in Arabidopsis autopolyploids and allopolyploids and their progenitors. Heredity 2012, 108, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Hodgkin, T.; Lyon, G.D.; Dickinson, H.G. Recognition in flowering plants: A comparison of the Brassica self-incompatibility system and plant pathogen interactions. New Phytol. 2010, 110, 557–569. [Google Scholar] [CrossRef]
- Sanabria, N.; Goring, D.; Nürnberger, T.; Dubery, I. Self/Nonself Perception and Recognition Mechanisms in Plants: A Comparison of Self-Incompatibility and Innate Immunity. New Phytol. 2008, 178, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, J.B. Recognition and rejection of self in plant self-incompatibility: Comparisons to animal histocompatibility. Trends Immunol. 2005, 26, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, R.; Gaude, T. Endocytosis and endosomal regulation of the S-receptor kinase during the self-incompatibility response in Brassica oleracea. Plant Cell 2009, 21, 2107–2117. [Google Scholar] [CrossRef] [PubMed]
- Muers, M. Gene expression: Transcriptome to proteome and back to genome. Nat. Rev. Genet. 2011, 12, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Odland, M.L.; Noll, C.J. The utilization of cross-compatibility and self-incompatibility in the production of F1 hybrid cabbage. Proc. Am. Soc. Horticult. Sci. 1950, 55, 391–402. [Google Scholar]
- Liu, H.L.; Fu, T.D.; Yang, X.N.; Wu, J.S. Preliminary report on the breeding of the maintainer lines and restoring lines of self-incompatible lines in Brassica napus. J. Huazhong Agric. Univ. 1981, 3, 9–28. [Google Scholar]
- Wang, C.; Zhang, L.; Yuan, M.; Ge, Y.; Liu, Y.; Fan, J.; Ruan, Y.; Cui, Z.; Tong, S.; Zhang, S. The microfilament cytoskeleton plays a vital role in salt and osmotic stress tolerance in Arabidopsis. Plant Biol. 2010, 12, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, J.; Yuan, M. Salt tolerance requires cortical microtubule reorganization in Arabidopsis. Plant Cell Physiol. 2007, 48, 1534–1547. [Google Scholar] [CrossRef] [PubMed]
- Iwano, M.; Shiba, H.; Matoba, K.; Miwa, T.; Funato, M.; Entani, T.; Nakayama, P.; Shimosato, H.; Takaoka, A.; Isogai, A.; et al. Actin dynamics in papilla cells of Brassica rapa during self- and cross-pollination. Plant Physiol. 2007, 144, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Dearnaley, J.D.W.; Clark, K.M.; Heath, I.B.; Lew, R.R.; Goring, D.R. Neither compatible nor self-incompatible pollinations of Brassica napus involve reorganization of the papillar cytoskeleton. New Phytol. 1999, 141, 199–207. [Google Scholar] [CrossRef]
- Safavian, D.; Goring, D.R. Secretory Activity Is Rapidly Induced in Stigmatic Papillae by Compatible Pollen, but Inhibited for Self-Incompatible Pollen in the Brassicaceae. PLoS ONE 2013, 8, e84286. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Sopory, S.K. Glyoxalase I from Brassica juncea: Molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress. Plant J. 1999, 17, 385–395. [Google Scholar] [CrossRef]
- Meng, M.; Geisler, M.; Johansson, H.; Harholt, J.; Scheller, H.V.; Mellerowicz, E.J.; Kleczkowski, L.A. UDP-glucose pyrophosphorylase is not rate limiting, but is essential in Arabidopsis. Plant Cell Physiol. 2009, 50, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.; Takeda, S.; Nick, P. Life and death under salt stress: Same players, different timing? J. Exp. Bot. 2014, 65, 2963–2979. [Google Scholar] [CrossRef] [PubMed]
- Sun-Wada, G.H.; Wada, Y. Role of vacuolar-type proton ATPase in signal transduction. Biochim. Biophys. Acta 2015, 1847, 1166–1172. [Google Scholar] [CrossRef] [PubMed]
- Tschochner, H.; Hurt, E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003, 13, 255–263. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; Nasrallah, J.B.; Nasrallah, M.E. Pollen–pistil interactions and developmental regulation of pollen tube growth in Arabidopsis. Development 1994, 120, 3405–3418. [Google Scholar]
- Elleman, C.J.; Franklin-Tong, V.; Dickinson, H.G. Pollination in species with dry stigmas: The nature of the early stigmatic response and the pathway taken by pollen tubes. New Phytol. 1992, 121, 413–424. [Google Scholar] [CrossRef]
- Tian, M.; Xie, Q. Non-26S proteasome proteolytic role of ubiquitin in plant endocytosis and endosomal trafficking(F.). J. Integr. Plant Biol. 2013, 55, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Estelle, M. The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 2010, 61, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Litkowski, P.E.; Taylor, A.A.; Lin, Y.; Snider, B.J.; Moulder, K.L. A role for the ubiquitin-proteasome system in activity-dependent presynaptic silencing. J. Neurosci. 2010, 30, 1798–1809. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M.; Franke, W.; Kleinschmidt, J.A. Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J. Biol. Chem. 1994, 269, 7709–7718. [Google Scholar] [PubMed]
- Robatzek, S.; Chinchilla, D.; Boller, T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genome Res. 2006, 20, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Geldner, N.; Hyman, D.L.; Wang, X.; Schumacher, K.; Chory, J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 2007, 21, 1598–1602. [Google Scholar] [CrossRef] [PubMed]
- Ungewickell, E.; Ungewickell, H.; Holstein, S.E.H.; Lindner, R.; Prasad, K.; Barouch, W.; Martini, B.; Greene, L.E.; Eisenberg, E. Role of auxilin in uncoating clathrin-coated vesicles. Nature 1995, 378, 632–635. [Google Scholar] [CrossRef] [PubMed]
- Schlossman, D.M.; Schmid, S.; Braell, W.A.; Rothman, J.E. An enzyme that removes clathrin coats: Purification of an uncoating ATPase. J. Cell Biol. 1984, 99, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Lam, B.C.-H.; Sage, T.L.; Bianchi, F.; Blumwald, E. Role of SH3 domain–containing proteins in clathrinmediated vesicle trafficking in Arabidopsis. Plant Cell 2001, 13, 2499–2512. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, J.; Maia, M.; Sousa Silva, M.; Figueiredo, A. Grapevine Subtilase Family: Update on New Sequences and Nomenclature Proposal. Front. Plant Sci. 2017, 8, 716. [Google Scholar] [CrossRef] [PubMed]
- Schaller, A.; Stintzi, A.; Graff, L. Subtilases—Versatile tools for protein turnover, plant development, and interactions with the environment. Physiol. Plant. 2012, 145, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Aderem, A.U.; David, M. Mechanisms of phagocytosis in macrophages. Ann. Rev. Immunol. 1999, 17, 593–623. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, J.J.; Brenner, M.B.; Thomas, D.Y.; Williams, D.B. Calnexin: A membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem. Sci. 1994, 19, 124–128. [Google Scholar] [CrossRef]
- Persson, S.; Rosenquist, M.; Svensson, K.; Galvao, R.; Boss, W.F.; Sommarin, M. Phylogenetic analyses and expression studies reveal two distinct groups of calreticulin isoforms in higher plants. Plant Physiol. 2003, 133, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Doucet, J.; Lee, H.K.; Goring, D.R. Pollen Acceptance or Rejection: A Tale of Two Pathways. Trends Plant Sci. 2016, 21, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Iwano, M.; Ito, K.; Fujii, S.; Kakita, M.; Asano-Shimosato, H.; Igarashi, M.; Kaothien-Nakayama, P.; Entani, T.; Kanatani, A.; Takehisa, M.; et al. Calcium signalling mediates self-incompatibility response in the Brassicaceae. Nat. Plants 2015, 1, 15128. [Google Scholar] [CrossRef] [PubMed]
- Ridley, B.L.; O’Neill, M.A.; Mohnen, D. Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 2001, 57, 929–967. [Google Scholar] [CrossRef]
- Showalter, A.M.; Keppler, B.; Lichtenberg, J.; Gu, D.; Welch, L.R. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 2010, 153, 485–513. [Google Scholar] [CrossRef] [PubMed]
- Kieliszewski, M.J.; Lamport, D.T. Extensin: Repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 1994, 5, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Cannon, M.C.; Terneus, K.; Hall, Q.; Tan, L.; Wang, Y.; Wegenhart, B.L.; Chen, L.; Lamport, D.T.; Chen, Y.; Kieliszewski, M.J. Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 2008, 105, 2226–2231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merkouropoulos, G.; Barnett, D.C.; Shirsat, A.H. The Arabidopsis extensin gene is developmentally regulated, is induced by wounding, methyl jasmonate, abscisic and salicylic acid, and codes for a protein with unusual motifs. Planta 1999, 208, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Kobe, B.; Deisenhofer, J. The leucine-rich repeat: A versatile binding motif. Trends Biochem. Sci. 1994, 19, 415–421. [Google Scholar] [CrossRef]
- Jaillais, Y.; Belkhadir, Y.; Balsemao-Pires, E.; Dangl, J.L.; Chory, J. Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc. Natl. Acad. Sci. USA 2011, 108, 8503–8507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, C.; Zhou, G.; Ma, C.; Zhai, W.; Zhang, T.; Liu, Z.; Yang, Y.; Wu, M.; Yue, Y.; Duan, Z.; et al. Helitron-like transposons contributed to the mating system transition from out-crossing to self-fertilizing in polyploid Brassica napus L. Sci. Rep. 2016, 6, 337–355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ma, C.; Fu, T.; Li, Y.; Wang, T.; Chen, Q.; Tu, J.; Shen, J. Development of SCAR markers linked to self-incompatibility in Brassica napus L. Mol. Breed. 2008, 21, 305–315. [Google Scholar] [CrossRef]
- Gao, C.; Ma, C.; Zhang, X.; Li, F.; Zhang, J.; Zhai, W.; Wang, Y.; Tu, J.; Shen, J.; Fu, T. The genetic characterization of self-incompatibility in a Brassica napus line with promising breeding potential. Mol. Breed. 2013, 31, 485–493. [Google Scholar] [CrossRef]
- Zhai, W.; Zhang, J.; Yang, Y.; Ma, C.; Liu, Z.; Gao, C.; Zhou, G.; Tu, J.; Shen, J.; Fu, T. Gene expression and genetic analysis reveal diverse causes of recessive self-compatibility in Brassica napus L. BMC Genom. 2014, 15, 1037. [Google Scholar] [CrossRef] [PubMed]







| Categories of Proteins | Number of Proteins | Pathway | Pathway ID | p-Value |
|---|---|---|---|---|
| PC/UP unique DAPs | 194 | Photosynthesis | ko00195 | 0.0023 |
| Amino sugar and nucleotide sugar metabolism | ko00520 | 0.0028 | ||
| Proteasome | ko03050 | 0.0036 | ||
| Metabolic pathways | ko01100 | 0.0063 | ||
| Cyanoamino acid metabolism | ko00460 | 0.0066 | ||
| Oxidative phosphorylation | ko00190 | 0.019 | ||
| Carbon fixation in photosynthetic organisms | ko00710 | 0.0224 | ||
| PI/UP unique DAPs | 71 | Inositol phosphate metabolism | ko00562 | 0.042 |
| Biosynthesis of secondary metabolites | ko01110 | 0.0457 | ||
| NA/UP unique DAPs | 134 | Ascorbate and aldarate metabolism | ko00053 | 0.0094 |
| Ribosome | ko03010 | 0.0099 | ||
| Plant–pathogen interaction | ko04626 | 0.0213 | ||
| Phagosome | ko04145 | 0.0259 | ||
| PC/UP_NA/UP intersection DAPs | 94 | Ribosome | ko03010 | 5.46 × 10−13 |
| Phagosome | ko04145 | 0.0202 | ||
| PC/UP_PI/UP intersection DAPs | 39 | Glyoxylate and dicarboxylate metabolism | ko00630 | 0.0133 |
| Alanine, aspartate, and glutamate metabolism | ko00250 | 0.0434 | ||
| PI/UP_NA/UP intersection DAPs | 17 | Glycine, serine, and threonine metabolism | ko00260 | 0.0177 |
| Biosynthesis of amino acids | ko01230 | 0.0273 | ||
| Biotin metabolism | ko00780 | 0.0417 | ||
| Metabolic pathways | ko01100 | 0.0478 | ||
| PC/UP_PI/UP_NA/UP intersection DAPs | 62 | Oxidative phosphorylation | ko00190 | 0.0129 |
| Photosynthesis | ko00195 | 0.0138 | ||
| Valine, leucine, and isoleucine degradation | ko00280 | 0.0138 | ||
| Plant–pathogen interaction | ko04626 | 0.0223 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Liu, Z.; Zhang, T.; Zhou, G.; Duan, Z.; Li, B.; Dou, S.; Liang, X.; Tu, J.; Shen, J.; et al. Mechanism of Salt-Induced Self-Compatibility Dissected by Comparative Proteomic Analysis in Brassica napus L. Int. J. Mol. Sci. 2018, 19, 1652. https://doi.org/10.3390/ijms19061652
Yang Y, Liu Z, Zhang T, Zhou G, Duan Z, Li B, Dou S, Liang X, Tu J, Shen J, et al. Mechanism of Salt-Induced Self-Compatibility Dissected by Comparative Proteomic Analysis in Brassica napus L. International Journal of Molecular Sciences. 2018; 19(6):1652. https://doi.org/10.3390/ijms19061652
Chicago/Turabian StyleYang, Yong, Zhiquan Liu, Tong Zhang, Guilong Zhou, Zhiqiang Duan, Bing Li, Shengwei Dou, Xiaomei Liang, Jinxing Tu, Jinxiong Shen, and et al. 2018. "Mechanism of Salt-Induced Self-Compatibility Dissected by Comparative Proteomic Analysis in Brassica napus L." International Journal of Molecular Sciences 19, no. 6: 1652. https://doi.org/10.3390/ijms19061652
APA StyleYang, Y., Liu, Z., Zhang, T., Zhou, G., Duan, Z., Li, B., Dou, S., Liang, X., Tu, J., Shen, J., Yi, B., Fu, T., Dai, C., & Ma, C. (2018). Mechanism of Salt-Induced Self-Compatibility Dissected by Comparative Proteomic Analysis in Brassica napus L. International Journal of Molecular Sciences, 19(6), 1652. https://doi.org/10.3390/ijms19061652