Interaction of Nevirapine with the Peptide Binding Groove of HLA-DRB1*01:01 and Its Effect on the Conformation of HLA-Peptide Complex
Abstract
:1. Introduction
2. Results
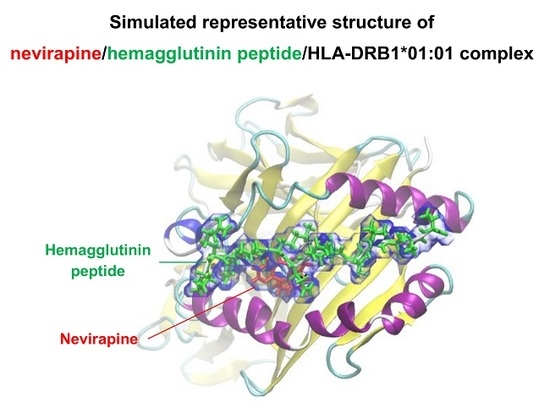
2.1. Docking Simulations
2.2. Molecular Dynamics (MD) Simulations
2.3. HLA Class II Competitive Assay
3. Discussion
4. Materials and Methods
4.1. Docking Simulations
4.2. MD Simulations
4.3. HLA Class II Competitive Assay
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| HLA | human leukocyte antigen |
| HA | hemagglutinin |
| HSR | hypersensitivity reaction |
| CYP | cytochrome P450 |
| SJS | Stevens-Johnson syndrome |
| TEN | toxic epidermal necrolysis |
| MD | molecular dynamics |
| GBVI/WSA_dG | generalized-Born volume integral/weighted surface area |
| RMSD | root mean square deviation |
| RMSF | root mean square fluctuation |
| SD | standard deviation |
| TT | tetanus toxoid |
| MBP | myelin basic protein |
| LC-MS/MS | liquid chromatography-tandem mass spectrometry |
| DMSO | dimethyl sulfoxide |
| CBD | chronic beryllium disease |
| TCR | T-cell receptor |
References
- Dieterich, D.T.; Robinson, P.A.; Love, J.; Stern, J.O. Drug-induced liver injury associated with the use of nonnucleoside reverse-transcriptase inhibitors. Clin. Infect. Dis. 2004, 38 (Suppl. 2), S80–S89. [Google Scholar] [CrossRef] [PubMed]
- Wit, F.W.; Kesselring, A.M.; Gras, L.; Richter, C.; van der Ende, M.E.; Brinkman, K.; Lange, J.M.; de Wolf, F.; Reiss, P. Discontinuation of nevirapine because of hypersensitivity reactions in patients with prior treatment experience, compared with treatment-naive patients: The ATHENA cohort study. Clin. Infect. Dis. 2008, 46, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Stern, J.O.; Robinson, P.A.; Love, J.; Lanes, S.; Imperiale, M.S.; Mayers, D.L. A comprehensive hepatic safety analysis of nevirapine in different populations of HIV infected patients. J. Acquir. Immune Defic. Syndr. 2003, 34 (Suppl. 1), S21–S33. [Google Scholar] [CrossRef] [PubMed]
- Keane, N.M.; Pavlos, R.K.; McKinnon, E.; Lucas, A.; Rive, C.; Blyth, C.C.; Dunn, D.; Lucas, M.; Mallal, S.; Phillips, E. HLA Class I restricted CD8+ and Class II restricted CD4+ T cells are implicated in the pathogenesis of nevirapine hypersensitivity. AIDS 2014, 28, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.M.; Nolan, D.; James, I.; Cameron, P.; Keller, J.; Moore, C.; Phillips, E.; Christiansen, F.T.; Mallal, S. Predisposition to nevirapine hypersensitivity associated with HLA-DRB1*0101 and abrogated by low CD4 T-cell counts. AIDS 2005, 19, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Guo, S.; Hall, D.; Cammett, A.M.; Jayadev, S.; Distel, M.; Storfer, S.; Huang, Z.; Mootsikapun, P.; Ruxrungtham, K.; et al. Toxicogenomics of nevirapine-associated cutaneous and hepatic adverse events among populations of African, Asian, and European descent. AIDS 2011, 25, 1271–1280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carr, D.F.; Chaponda, M.; Jorgensen, A.L.; Castro, E.C.; van Oosterhout, J.J.; Khoo, S.H.; Lalloo, D.G.; Heyderman, R.S.; Alfirevic, A.; Pirmohamed, M. Association of human leukocyte antigen alleles and nevirapine hypersensitivity in a Malawian HIV-infected population. Clin. Infect. Dis. 2013, 56, 1330–1339. [Google Scholar] [CrossRef] [PubMed]
- Carr, D.F.; Bourgeois, S.; Chaponda, M.; Takeshita, L.Y.; Morris, A.P.; Castro, E.M.; Alfirevic, A.; Jones, A.R.; Rigden, D.J.; Haldenby, S.; et al. Genome-wide association study of nevirapine hypersensitivity in a sub-Saharan African HIV-infected population. J. Antimicrob. Chemother. 2017, 72, 1152–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penzak, S.R.; Kabuye, G.; Mugyenyi, P.; Mbamanya, F.; Natarajan, V.; Alfaro, R.M.; Kityo, C.; Formentini, E.; Masur, H. Cytochrome P450 2B6 (CYP2B6) G516T influences nevirapine plasma concentrations in HIV-infected patients in Uganda. HIV Med. 2007, 8, 86–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, D.W.; Bartlett, J.A.; Andersen, J.W.; Sanne, I.; Wilkinson, G.R.; Hinkle, J.; Rousseau, F.; Ingram, C.D.; Shaw, A.; Lederman, M.M.; et al. Pharmacogenetics of nevirapine-associated hepatotoxicity: An Adult AIDS Clinical Trials Group collaboration. Clin. Infect. Dis. 2006, 43, 783–786. [Google Scholar] [CrossRef] [PubMed]
- Chessman, D.; Kostenko, L.; Lethborg, T.; Purcell, A.W.; Williamson, N.A.; Chen, Z.; Kjer-Nielsen, L.; Mifsud, N.A.; Tait, B.D.; Holdsworth, R.; et al. Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity. Immunity 2008, 28, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Somkrua, R.; Eickman, E.E.; Saokaew, S.; Lohitnavy, M.; Chaiyakunapruk, N. Association of HLA-B*5801 allele and allopurinol-induced Stevens Johnson syndrome and toxic epidermal necrolysis: A systematic review and meta-analysis. BMC Med. Genet. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Pavlos, R.; McKinnon, E.J.; Ostrov, D.A.; Peters, B.; Buus, S.; Koelle, D.; Chopra, A.; Schutte, R.; Rive, C.; Redwood, A.; et al. Shared peptide binding of HLA Class I and II alleles associate with cutaneous nevirapine hypersensitivity and identify novel risk alleles. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Cornejo Castro, E.M.; Carr, D.F.; Jorgensen, A.L.; Alfirevic, A.; Pirmohamed, M. HLA-allelotype associations with nevirapine-induced hypersensitivity reactions and hepatotoxicity: A systematic review of the literature and meta-analysis. Pharmacogenet. Genom. 2015, 25, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Umapathy, S.; Pawar, A.; Bajpai, S.; Pazare, A.R.; Ghosh, K. HLA involvement in nevirapine-induced dermatological reaction in antiretroviral-treated HIV-1 patients. J. Pharmacol. Pharmacother. 2011, 2, 114–115. [Google Scholar] [CrossRef] [PubMed]
- Pavlos, R.; Mallal, S.; Phillips, E. HLA and pharmacogenetics of drug hypersensitivity. Pharmacogenomics 2012, 13, 1285–1306. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, M.; Hagihara, K.; Okudaira, N.; Izumi, T. The possible mechanism of idiosyncratic lapatinib-induced liver injury in patients carrying human leukocyte antigen-DRB1*07:01. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, M.; Hagihara, K.; Abe, K.; Ando, O.; Hirayama, N. In Silico and In Vitro analysis of interaction between ximelagatran and human leukocyte antigen (HLA)-DRB1*07:01. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Isogai, H.; Hirayama, N. In Silico analysis of interactions between nevirapine-related compounds, HLA-B*14:02 and T-cell receptor. Chem-Bio Inf. J. 2016, 16, 9–12. [Google Scholar]
- Rabinowitz, J.D.; Vrljic, M.; Kasson, P.M.; Liang, M.N.; Busch, R.; Boniface, J.J.; Davis, M.M.; McConnell, H.M. Formation of a Highly peptide-receptive state of class II MHC. Immunity 1998, 9, 699–709. [Google Scholar] [CrossRef]
- Osabe, M.; Tohkin, M.; Hirayama, N. In Silico analysis of interactions between HLA-B*58:01 and allopurinol-related compounds. Chem-Bio Inf. J. 2016, 16, 1–4. [Google Scholar] [CrossRef]
- Illing, P.T.; Vivian, J.P.; Dudek, N.L.; Kostenko, L.; Chen, Z.; Bharadwaj, M.; Miles, J.J.; Kjer-Nielsen, L.; Gras, S.; Williamson, N.A.; et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 2012, 486, 554–558. [Google Scholar] [CrossRef] [PubMed]
- Clayton, G.M.; Wang, Y.; Crawford, F.; Novikov, A.; Wimberly, B.T.; Kieft, J.S.; Falta, M.T.; Bowerman, N.A.; Marrack, P.; Fontenot, A.P.; et al. Structural basis of chronic beryllium disease: Linking allergic hypersensitivity and autoimmunity. Cell 2014, 158, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Petukh, M.; Wu, B.; Stefl, S.; Smith, N.; Hyde-Volpe, D.; Wang, L.; Alexov, E. Chronic beryllium disease: Revealing the role of beryllium ion and small peptides binding to HLA-DP2. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, F.C.; Koetzle, T.F.; Williams, G.J.; Meyer, E.F., Jr.; Brice, M.D.; Rodgers, J.R.; Kennard, O.; Shimanouchi, T.; Tasumi, M. The protein data bank: A computer-based archival file for macromolecular structures. J. Mol. Biol. 1977, 112, 535–542. [Google Scholar] [CrossRef]
- Goto, J.; Kataoka, R.; Muta, H.; Hirayama, N. ASEDock-docking based on alpha spheres and excluded volumes. J. Chem. Inf. Model. 2008, 48, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, C.R.; Williams, C.I.; Labute, P.; Spraggs, C.F. Variability in docking success rates due to dataset preparation. J. Comput. Aided Mol. Des. 2012, 26, 775–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chemical Computing Group Inc. Molecular Operating Environment (MOE), 2013.08; Chemical Computing Group Inc.: Montreal, QC, Canada, 2017. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Steere, A.C.; Klitz, W.; Drouin, E.E.; Falk, B.A.; Kwok, W.W.; Nepom, G.T.; Baxter-Lowe, L.A. Antibiotic-refractory Lyme arthritis is associated with HLA-DR molecules that bind a Borrelia burgdorferi peptide. J. Exp. Med. 2006, 203, 961–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Hemagglutinin Peptide | Nevirapine | Energies (kcal/mol) | RMSF (Å) | Inter-Helical Distance (Å) | Slope of Inter-Helical Distance (Å/ns) |
|---|---|---|---|---|---|
| - | − | −106715 ± 309.9 | 1.0 ± 0.4 | 13.0 ± 0.4 | −0.12 |
| - | + | −106370 ± 533.5 | 0.8 ± 0.3 | 13.1 ± 0.4 | −0.14 |
| Frame 2 | − | −111392 ± 579.8 | 0.9 ± 0.4 | 12.7 ± 0.2 | −0.16 |
| Frame 3 | − | −106078 ± 323.1 | 0.9 ± 0.3 | 14.2 ± 0.3 | 0.04 |
| Frame 2 | + | −120550 ± 243.6 | 0.8 ± 0.3 | 13.5 ± 0.2 | 0.02 |
| Frame 3 | + | −112166 ± 230.5 | 0.9 ± 0.3 | 12.6 ± 0.2 | −0.13 |
| HLA Allele | DRB1*01:01 | DRB1*07:01 | DRB1*15:01 | |
|---|---|---|---|---|
| Concentration of nevirapine (μM) | 1000 | 263.9 ± 15.5 # | 62.6 ± 3.9 | 101.3 ± 14.9 |
| 200 | 99.7 ± 11.3 | 110.4 ± 5.5 | 94.6 ± 10.6 | |
| 40 | 106.2 ± 6.5 | 76.9 ± 4.6 | 93 ± 10.4 | |
| 8 | 101.9 ± 8.1 | 132.2 ± 64 | 98.9 ± 3.4 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirasawa, M.; Hagihara, K.; Abe, K.; Ando, O.; Hirayama, N. Interaction of Nevirapine with the Peptide Binding Groove of HLA-DRB1*01:01 and Its Effect on the Conformation of HLA-Peptide Complex. Int. J. Mol. Sci. 2018, 19, 1660. https://doi.org/10.3390/ijms19061660
Hirasawa M, Hagihara K, Abe K, Ando O, Hirayama N. Interaction of Nevirapine with the Peptide Binding Groove of HLA-DRB1*01:01 and Its Effect on the Conformation of HLA-Peptide Complex. International Journal of Molecular Sciences. 2018; 19(6):1660. https://doi.org/10.3390/ijms19061660
Chicago/Turabian StyleHirasawa, Makoto, Katsunobu Hagihara, Koji Abe, Osamu Ando, and Noriaki Hirayama. 2018. "Interaction of Nevirapine with the Peptide Binding Groove of HLA-DRB1*01:01 and Its Effect on the Conformation of HLA-Peptide Complex" International Journal of Molecular Sciences 19, no. 6: 1660. https://doi.org/10.3390/ijms19061660
APA StyleHirasawa, M., Hagihara, K., Abe, K., Ando, O., & Hirayama, N. (2018). Interaction of Nevirapine with the Peptide Binding Groove of HLA-DRB1*01:01 and Its Effect on the Conformation of HLA-Peptide Complex. International Journal of Molecular Sciences, 19(6), 1660. https://doi.org/10.3390/ijms19061660