Comparative Transcriptomic Analysis of Immune-Related Gene Expression in Duck Embryo Fibroblasts Following Duck Tembusu Virus Infection
Abstract
:1. Introduction
2. Results
2.1. DTMUV Infection Induces Cytopathic Effects (CPEs) and Reduces Cell Viability
2.2. Transcriptome Sequencing Data
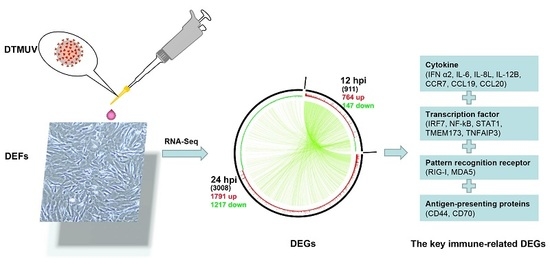
2.3. Identification and Analysis of DEGs
2.4. GO and KEGG Enrichment Analysis
2.5. Signaling Pathways in the Immune System
2.6. Expression of Key DEGs in Immune-Related Pathways
2.7. Validation of the Transcriptomic Data by Quantitative Real-Time (qRT)-PCR
2.8. Determination of Cytokine Levels in Culture Supernatant
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Virus Infection
4.2. Cell Viability Detection by WST-1 Assay
4.3. DEFs Genomic RNA Extraction, Library Construction and Illumina Sequencing
4.4. Transcriptome Data Analysis
4.5. Differential Expression Analysis
4.6. Validation of RNA-Seq Data by qRT-PCR
4.7. Determination of Cytokine Levels in Culture Supernatants Using Enzyme-Linked Immunoassay (ELISA)
5. Conclusions
Reference
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lei, W.; Guo, X.; Fu, S.; Feng, Y.; Tao, X.; Gao, X.; Song, J.; Yang, Z.; Zhou, H.; Liang, G. The genetic characteristics and evolution of Tembusu virus. Vet. Microbiol. 2017, 201, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Yu, X.; Yang, G.; Tang, Y.; Diao, Y. A Novel Diagnostic Method to Detect Duck Tembusu Virus: A Colloidal Gold-Based Immunochromatographic Assay. Front. Microbiol. 2018, 9, 1001. [Google Scholar] [CrossRef] [PubMed]
- Ti, J.; Zhang, L.; Li, Z.; Zhao, D.; Zhang, Y.; Li, F.; Diao, Y. Effect of age and inoculation route on the infection of duck Tembusu virus in Goslings. Vet. Microbiol. 2015, 181, 190–197. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, A.; Chen, S.; Wu, Z.; Zhang, J.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Yang, Q. Differential immune-related gene expression in the spleens of duck Tembusu virus-infected goslings. Vet. Microbiol. 2017, 212, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Chen, C.; Huang, Y.; Cheng, L.; Fu, Q.; Wan, C.; Shi, S.; Chen, H.; Liu, W. Comparative analysis of transcriptional profiles of retinoic-acid-induced gene I-like receptors and interferons in seven tissues from ducks infected with avian Tembusu virus. Arch. Virol. 2016, 161, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Diao, Y.; Yu, C.; Gao, X.; Ju, X.; Xue, C.; Liu, X.; Ge, P.; Qu, J.; Zhang, D. Characterization of a Tembusu virus isolated from naturally infected house sparrows (Passer domesticus) in Northern China. Transbound. Emerg. Dis. 2013, 60, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Zhang, L.; Wang, Y.; Yu, X.; Tian, K.; Su, W.; Han, B.; Su, J. Duck Tembusu virus exhibits neurovirulence in BALB/c mice. Virol. J. 2013, 10, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ti, J.; Zhang, M.; Li, Z.; Li, X.; Diao, Y. Duck Tembusu Virus Exhibits Pathogenicity to Kunming Mice by Intracerebral Inoculation. Front. Microbiol. 2016, 7, 190. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Lu, H.; Li, S.; Moureau, G.; Deng, Y.Q.; Wang, Y.; Zhang, L.; Jiang, T.; de Lamballerie, X.; Qin, C.F. Genomic and antigenic characterization of the newly emerging Chinese duck egg-drop syndrome flavivirus: Genomic comparison with Tembusu and Sitiawan viruses. J. Gen. Virol. 2012, 93, 2158–2170. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.J.; Liu, L.; Li, X.F.; Ye, Q.; Deng, Y.Q.; Qin, E.D.; Qin, C.F. In vitro and in vivo characterization of chimeric duck Tembusu virus based on Japanese encephalitis live vaccine strain SA14-14-2. J. Gen. Virol. 2016, 97, 1551–1556. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Gao, X.; Diao, Y.; Feng, Q.; Chen, H.; Liu, X.; Ge, P.; Yu, C. Tembusu virus in human, China. Transbound. Emerg. Dis. 2013, 60, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Liu, F.; Chen, S.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Sun, K.; Wu, Y.; Chen, X.; et al. Transcriptome Analysis and Identification of Differentially Expressed Transcripts of Immune-Related Genes in Spleen of Gosling and Adult Goose. Int. J. Mol. Sci. 2015, 16, 22904–22926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, F.L.; Liu, X.; Han, Z.X.; Shao, Y.H.; Kong, X.G.; Liu, S.W. Transcriptome analysis of chicken kidney tissues following coronavirus avian infectious bronchitis virus infection. BMC Genom. 2013, 14, 743. [Google Scholar] [CrossRef] [PubMed]
- Jagya, N.; Varma, S.P.K.; Thakral, D.; Joshi, P.; Duragapal, H.; Panda, S.K. RNA-Seq Based Transcriptome Analysis of Hepatitis E Virus (HEV) and Hepatitis B Virus (HBV) Replicon Transfected Huh-7 Cells. PLoS ONE 2014, 9, e87835. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, K.E.; Nalpas, N.C.; Rue-Albrecht, K.; Browne, J.A.; Magee, D.A.; Killick, K.E.; Park, S.D.; Hokamp, K.; Meade, K.G.; O’Farrelly, C.; et al. RNA-seq Transcriptional Profiling of Peripheral Blood Leukocytes from Cattle Infected with Mycobacterium bovis. Front. Immunol. 2014, 5, 396. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.K.; Li, S.C.; Qi, K.Z.; Xue, T.; Tu, J.; Zhou, X.H.; Liu, H.M. Application of RNA-Seq technology for screening differentially expressed genes in chicken small intestine cells affected by avian pathogenic Escherichia coli. Chin. J. Prev. Vet. Med. 2016, 38, 111–115. [Google Scholar]
- Menicucci, A.R.; Versteeg, K.; Woolsey, C.; Mire, C.E.; Geisbert, J.B.; Cross, R.W.; Agans, K.N.; Jankeel, A.; Geisbert, T.W.; Messaoudi, I. Transcriptome Analysis of Circulating Immune Cell Subsets Highlight the Role of Monocytes in Zaire Ebola Virus Makona Pathogenesis. Front. Immunol. 2017, 8, 1372. [Google Scholar] [CrossRef] [PubMed]
- Preusse, M.; Schughart, K.; Pessler, F. Host Genetic Background Strongly Affects Pulmonary microRNA Expression before and during Influenza A Virus Infection. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; He, L.; Luo, L.; Huang, R.; Liao, L.; Li, Y.; Zhu, Z.; Wang, Y. Transcriptomics Sequencing Provides Insights into Understanding the Mechanism of Grass Carp Reovirus Infection. Int. J. Mol. Sci. 2018, 19, 488. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Gao, Q.; Tang, B.; Sun, P.; Han, K.; Huang, W. Transcriptome and analysis on the complement and coagulation cascades pathway of large yellow croaker (Larimichthys crocea) to ciliate ectoparasite Cryptocaryon irritans infection. Fish Shellfish Immunol. 2016, 50, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Lv, C.; Yue, R.; Shi, Y.; Wei, L.; Chai, T.; Liu, S. Effect of age on the pathogenesis of duck tembusu virus in Cherry Valley ducks. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.Q.; Lin, W.C.; Wang, Z.X.; Zhang, K.; Yan, Z.Q.; Zhou, Q.F.; Qin, J.P.; Xie, Q.M.; Bi, Y.Z.; Chen, F. Pathogenicity and genetic characterization of a duck Tembusu virus associated with egg-dropping in Muscovy ducks. Virus Res. 2016, 223, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Yeh, Y.T.; Chen, H.; Yu, C.; Gao, X.; Diao, Y. Comparison of four molecular assays for the detection of Tembusu virus. Avian Pathol. 2015, 44, 379–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Shi, Y.; Yan, D.; Li, X.; Yan, P.; Gao, X.; Zhang, Y.; Yu, L.; Ren, C.; Li, G.; et al. Development of a PCR-Based Reverse Genetics System for an Attenuated Duck Tembusu Virus Strain. PLoS ONE 2016, 11, e0156579. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Liu, Y.; Cheng, J.; Liu, Y.; Fan, W.; Cheng, Z.; Niu, X.; Liu, J. Liposomes containing recombinant E protein vaccine against duck Tembusu virus in ducks. Vaccine 2016, 34, 2157–2163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Z.; Zhang, Q.; Sun, M.; Li, S.; Su, W.; Hu, X.; He, W.; Su, J. Efficacy assessment of an inactivated Tembusu virus vaccine candidate in ducks. Res. Vet. Sci. 2017, 110, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, X.; Chen, H.; Ti, J.; Yang, G.; Zhang, L.; Lu, Y.; Diao, Y. Evidence of possible vertical transmission of Tembusu virus in ducks. Vet. Microbiol. 2015, 179, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Li, Z.; Tao, P. Evolutionary analysis of Tembusu virus: Evidence for the emergence of a dominant genotype. Infect. Genet. Evol. 2015, 32, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, T.; Song, D.; Huang, T.; Peng, Q.; Chen, Y.; Li, A.; Zhang, F.; Wu, Q.; Ye, Y.; Tang, Y. Whole-Genome Sequence of Duck Tembusu Virus Strain DTMUV/CH/2014, Isolated in China. Genome Announc. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Zhao, D.; Liu, Y.; Huang, X.; Yang, J.; Liu, Q.; An, F.; Li, Y. Design and evaluation of a polytope construct with multiple B and T epitopes against Tembusu virus infection in ducks. Res. Vet. Sci. 2016, 104, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Huang, X.; Liu, Y.; Han, K.; Zhang, J.; Yang, J.; Xie, X.; Li, Y. Domain I and II from newly emerging goose tembusu virus envelope protein functions as a dominant-negative inhibitor of virus infectivity. Res. Vet. Sci. 2015, 98, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, Y.; Li, R.; Liu, J.; Zhang, J.; Cai, Y.; Liu, S.; Chai, T.; Wei, L. Immune responses of ducks infected with duck Tembusu virus. Front. Microbiol. 2015, 6, 425. [Google Scholar] [CrossRef] [PubMed]
- Chazal, M.; Beauclair, G.; Gracias, S.; Najburg, V.; Simon-Loriere, E.; Tangy, F.; Komarova, A.V.; Jouvenet, N. RIG-I Recognizes the 5’ Region of Dengue and Zika Virus Genomes. Cell Rep. 2018, 24, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.M.; Fornek, J.; Crochet, N.; Bajwa, G.; Perwitasari, O.; Martinez-Sobrido, L.; Akira, S.; Gill, M.A.; Garcia-Sastre, A.; Katze, M.G.; et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008, 82, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Urcuqui-Inchima, S.; Cabrera, J.; Haenni, A.L. Interplay between dengue virus and Toll-like receptors, RIG-I/MDA5 and microRNAs: Implications for pathogenesis. Antivir. Res. 2017, 147, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Khandia, R.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Malik, Y.S.; Singh, R.K.; Chaicumpa, W. Modulation of Dengue/Zika Virus Pathogenicity by Antibody-Dependent Enhancement and Strategies to Protect Against Enhancement in Zika Virus Infection. Front. Immunol. 2018, 9, 597. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Xiao, F.; Hong, J.; Wang, K.; Liu, X.; Cai, D.; Fusco, D.N.; Zhao, L.; Jeong, S.W.; Brisac, C.; et al. EFTUD2 is a Novel Innate Immune Regulator Restricting Hepatitis C Virus Infection through the RIG-I/MDA5 Pathway. J. Virol. 2015, 89, 6608–6618. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamto, M.; Matsui, K.; Uematsu, S.; Jung, A. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Fredericksen, B.L.; Keller, B.C.; Fornek, J.; Katze, M.G.; Gale, M.J. Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 2008, 82, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Ye, H.Q.; Liu, S.Q.; Deng, C.L.; Li, X.D.; Shi, P.Y.; Zhang, B. West Nile Virus NS1 Antagonizes Interferon Beta Production by Targeting RIG-I and MDA5. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Livolsi, A.; Busuttil, V.; Imbert, V.; Abraham, R.T.; Peyron, J.F. Tyrosine phosphorylation-dependent activation of NF-κB Requirement for p56 LCK and ZAP-70 protein tyrosine kinases. Eur. J. Biochem. 2001, 268, 1508–1515. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Ni, L.; Wang, S.; Wang, K.; Lin, R.; Zheng, C. Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-kappaB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP. J. Virol. 2013, 87, 9788–9801. [Google Scholar] [CrossRef] [PubMed]
- Shahsavandi, S.; Ebrahimi, M.M.; Mohammadi, A.; Zarrin Lebas, N. Impact of chicken-origin cells on adaptation of a low pathogenic influenza virus. Cytotechnology 2013, 65, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.J.; Liu, G.J. Progress and Prospect of STAT1 in Tumorigenesis. Chin. J. Cell Biol. 2017, 39, 1612–1628. [Google Scholar]
- Adams, S.; Xing, Z.; Li, J.; Mendoza, K.; Perez, D.; Reed, K.; Cardona, C. The effect of avian influenza virus NS1 allele on virus replication and innate gene expression in avian cells. Mol. Immunol. 2013, 56, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xie, P.; Sun, M.; Xiang, B.; Kang, Y.; Gao, P.; Zhu, W.; Ning, Z.; Ren, T. S1PR1 expression correlates with inflammatory responses to Newcastle disease virus infection. Infect. Genet Evol. 2016, 37, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–11. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L. Trinity: Reconstructing a full-length transcriptome without a reference genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 50, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]










| Sample | Raw Reads | Clean Reads | Clean Data (%) | Q20 (%) | Q30 (%) | Total Mapped (%) |
|---|---|---|---|---|---|---|
| 12hD1 | 82,525,576 | 81,938,158 | 99.28 | 97.70 | 94.29 | 76.51 |
| 12hD2 | 82,444,204 | 81,830,302 | 99.25 | 97.66 | 94.24 | 77.84 |
| 12hD3 | 82,505,912 | 81,871,548 | 99.23 | 97.62 | 94.15 | 76.91 |
| 12hS1 | 81,086,676 | 80,390,534 | 99.14 | 97.70 | 94.28 | 69.68 |
| 12hS2 | 81,606,422 | 81,000,120 | 99.25 | 97.77 | 94.41 | 61.35 |
| 12hS3 | 84,368,222 | 83,680,924 | 99.18 | 97.72 | 94.30 | 65.83 |
| 24hD1 | 84,046,382 | 83,435,382 | 99.28 | 97.63 | 94.15 | 73.81 |
| 24hD2 | 85,114,912 | 84,487,666 | 99.26 | 97.57 | 94.03 | 71.89 |
| 24hD3 | 85,158,520 | 84,518,936 | 99.24 | 97.56 | 94.03 | 73.36 |
| 24hS1 | 84,904,016 | 83,892,832 | 98.80 | 97.70 | 94.24 | 46.64 |
| 24hS2 | 85,411,010 | 84,413,208 | 98.83 | 97.82 | 94.46 | 45.81 |
| 24hS3 | 84,786,930 | 83,734,226 | 98.75 | 97.68 | 94.17 | 45.10 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, G.; Lin, Y.; Tang, Y.; Diao, Y. Comparative Transcriptomic Analysis of Immune-Related Gene Expression in Duck Embryo Fibroblasts Following Duck Tembusu Virus Infection. Int. J. Mol. Sci. 2018, 19, 2328. https://doi.org/10.3390/ijms19082328
Yu G, Lin Y, Tang Y, Diao Y. Comparative Transcriptomic Analysis of Immune-Related Gene Expression in Duck Embryo Fibroblasts Following Duck Tembusu Virus Infection. International Journal of Molecular Sciences. 2018; 19(8):2328. https://doi.org/10.3390/ijms19082328
Chicago/Turabian StyleYu, Guanliu, Yun Lin, Yi Tang, and Youxiang Diao. 2018. "Comparative Transcriptomic Analysis of Immune-Related Gene Expression in Duck Embryo Fibroblasts Following Duck Tembusu Virus Infection" International Journal of Molecular Sciences 19, no. 8: 2328. https://doi.org/10.3390/ijms19082328
APA StyleYu, G., Lin, Y., Tang, Y., & Diao, Y. (2018). Comparative Transcriptomic Analysis of Immune-Related Gene Expression in Duck Embryo Fibroblasts Following Duck Tembusu Virus Infection. International Journal of Molecular Sciences, 19(8), 2328. https://doi.org/10.3390/ijms19082328