Exosomal Chaperones and miRNAs in Gliomagenesis: State-of-Art and Theranostics Perspectives
Abstract
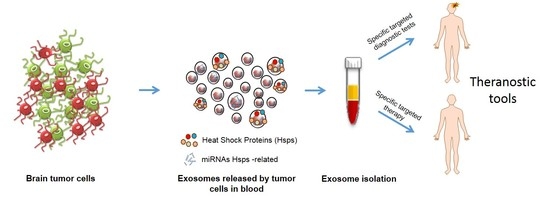
:1. Introduction
2. Gliomagenesis Overview and Clinical and Histopathological Classifications of Gliomas
3. Molecular Chaperones: Locales of Residence, Functions, and Roles during Tumorigenesis
4. miRNAs: Intracellular Localization, Functions, and Roles during Tumorigenesis
5. Exosomes: Nanovectors for Extracellular Chaperones and miRNAs
6. Chaperones, miRNAs, and Exosomes as Novel Molecular Target to Be Explored for Diagnosis and Prognosis Assessment of Gliomas
7. Potential Therapeutic Applications and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CNS | central nervous system |
| PNS | peripheral nervous system |
| NSCs | neural stem cells |
| GBM | glioblastoma multiforme |
| WHO | world health organization |
| ECM | extracellular matrix |
| EMT | epithelial to mesenchymal transition |
| MMPs | matrix metalloproteases |
| DWI | diffusion-weighted imaging |
| DTI | diffusion tensor imaging |
| DTT | diffusion tensor tractography |
| MRI | magnetic resonance imaging |
| MRS | magnetic resonance spectroscopy |
| PET | positron emission tomography |
| Hsp | heat shock protein |
| MVB | multivesicular bodies |
| ILVs | intraluminal vesicles |
References
- Ostrom, Q.T.; Gittleman, H.; Farah, P.; Ondracek, A.; Chen, Y.; Wolinsky, Y.; Stroup, N.E.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2006–2010. Neuro. Oncol. 2013, 15, ii1–ii56. [Google Scholar] [CrossRef] [PubMed]
- Caruso Bavisotto, C.; Cappello, F.; Macario, A.J.L.; Conway de Macario, E.; Logozzi, M.; Fais, S.; Campanella, C. Exosomal Hsp60: A potentially useful biomarker for diagnosis, assessing prognosis, and monitoring response to treatment. Expert Rev. Mol. Diagn. 2017, 17, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Fais, S.; O’Driscoll, L.; Borras, F.E.; Buzas, E.; Camussi, G.; Cappello, F.; Carvalho, J.; Cordeiro da Silva, A.; Del Portillo, H.; El Andaloussi, S.; et al. Evidence-Based Clinical Use of Nanoscale Extracellular Vesicles in Nanomedicine. ACS Nano 2016, 10, 3886–3899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iessi, E.; Logozzi, M.; Lugini, L.; Azzarito, T.; Federici, C.; Spugnini, E.P.; Mizzoni, D.; Di Raimo, R.; Angelini, D.F.; Battistini, L.; et al. Acridine Orange/exosomes increase the delivery and the effectiveness of Acridine Orange in human melanoma cells: A new prototype for theranostics of tumors. J. Enzym. Inhib. Med. Chem. 2017, 32, 648–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, D.L.; Johansson, C.B.; Wilbertz, J.; Veress, B.; Nilsson, E.; Karlström, H.; Lendahl, U.; Frisén, J. Generalized potential of adult neural stem cells. Science 2000, 288, 1660–1663. [Google Scholar] [CrossRef] [PubMed]
- Crocetti, E.; Trama, A.; Stiller, C.; Caldarella, A.; Soffietti, R.; Jaal, J.; Weber, D.C.; Ricardi, U.; Slowinski, J.; Brandes, A.; et al. Epidemiology of glial and non-glial brain tumours in Europe. Eur. J. Cancer 2012, 48, 1532–1542. [Google Scholar] [CrossRef] [PubMed]
- Alifieris, C.; Trafalis, D.T. Glioblastoma multiforme: Pathogenesis and treatment. Pharmacol. Ther. 2015, 152, 63–82. [Google Scholar] [CrossRef] [PubMed]
- De Robles, P.; Fiest, K.M.; Frolkis, A.D.; Pringsheim, T.; Atta, C.; St Germaine-Smith, C.; Day, L.; Lam, D.; Jette, N. The worldwide incidence and prevalence of primary brain tumors: A systematic review and meta-analysis. Neuro-Oncology 2015, 17, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Brada, M.; van den Bent, M.J.; Tonn, J.C.; Pentheroudakis, G.; ESMO Guidelines Working Group. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25, iii93–iii101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appin, C.L.; Brat, D.J. Molecular Genetics of Gliomas. Cancer J. 2014, 20, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Iser, I.C.; Pereira, M.B.; Lenz, G.; Wink, M.R. The Epithelial-to-Mesenchymal Transition-Like Process in Glioblastoma: An Updated Systematic Review and In Silico Investigation. Med. Res. Rev. 2017, 37, 271–313. [Google Scholar] [CrossRef] [PubMed]
- Carro, M.S.; Lim, W.K.; Alvarez, M.J.; Bollo, R.J.; Zhao, X.; Snyder, E.Y.; Sulman, E.P.; Anne, S.L.; Doetsch, F.; Colman, H.; et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 2010, 463, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, Y.; Biswas, A.; Mandal, M. Glioma progression through the prism of heat shock protein mediated extracellular matrix remodeling and epithelial to mesenchymal transition. Exp. Cell Res. 2017, 359, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Kesari, S. Malignant gliomas in adults. N. Engl. J. Med. 2008, 359, 492–507. [Google Scholar] [CrossRef] [PubMed]
- Czarnecka, A.M.; Campanella, C.; Zummo, G.; Cappello, F. Mitochondrial chaperones in cancer: From molecular biology to clinical diagnostics. Cancer Biol. Ther. 2006, 5, 714–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macario, A.J.L.; Conway de Macario, E. Sick chaperones, cellular stress, and disease. N. Engl. J. Med. 2005, 353, 1489–1501. [Google Scholar] [CrossRef] [PubMed]
- Macario, A.J.L.; Conway de Macario, E. Chaperonopathies and chaperonotherapy. FEBS Lett. 2007, 581, 3681–3688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macario, A.J.L.; Cappello, F.; Zummo, G.; Conway de Macario, E. Chaperonopathies of senescence and the scrambling of interactions between the chaperoning and the immune systems. Ann. N. Y. Acad. Sci. 2010, 1197, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Macario, A.J.L.; Conway de Macario, E.; Cappello, F. The Chaperonopathies: Diseases with Defective Molecular Chaperones; Springer: Dordrecht, The Netherlands, 2013. [Google Scholar]
- Macario, A.J.L.; Conway de Macario, E. Chaperone proteins and chaperonopathies. In Stress Physiology, Biochemistry, and Pathology. Handbook of Stress; Fink, G., Ed.; Elsevier: Amsterdam, The Netherlands, in press.
- Lindquist, S. The Heat-Shock Response. Annu. Rev. Biochem. 1986, 55, 1151–1191. [Google Scholar] [CrossRef] [PubMed]
- Cappello, F.; Marino Gammazza, A.; Palumbo Piccionello, A.; Campanella, C.; Pace, A.; Conway de Macario, E.; Macario, A.J.L. Hsp60 chaperonopathies and chaperonotherapy: Targets and agents. Expert Opin. Ther. Targets 2014, 18, 185–208. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.K.; Stevenson, M.A.; Murshid, A. Heat Shock Proteins, Autoimmunity, and Cancer Treatment. Autoimmune Dis. 2012, 2012, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, Y.; Torigoe, T.; Kukita, K.; Saito, K.; Okuya, K.; Kutomi, G.; Hirata, K.; Sato, N. Heat-shock proteins as endogenous ligands building a bridge between innate and adaptive immunity. Immunotherapy 2012, 4, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Fagone, P.; Di Rosa, M.; Palumbo, M.; De Gregorio, C.; Nicoletti, F.; Malaguarnera, L. Modulation of heat shock proteins during macrophage differentiation. Inflamm. Res. 2012, 61, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Rappa, F.; Farina, F.; Zummo, G.; David, S.; Campanella, C.; Carini, F.; Tomasello, G.; Damiani, P.; Cappello, F.; Conway de Macario, E.; et al. Hsp-molecular chaperones in cancer biogenesis and tumor therapy: An overview. Anticancer Res. 2012, 32, 5139–5150. [Google Scholar] [PubMed]
- Kast, R.E.; Boockvar, J.A.; Brüning, A.; Cappello, F.; Chang, W.-W.; Cvek, B.; Dou, Q.P.; Duenas-Gonzalez, A.; Efferth, T.; Focosi, D.; et al. A conceptually new treatment approach for relapsed glioblastoma: Coordinated undermining of survival paths with nine repurposed drugs (CUSP9) by the International Initiative for Accelerated Improvement of Glioblastoma Care. Oncotarget 2013, 4, 502–530. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. Challenging the dogma: The hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 2003, 25, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Chang, H.Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013, 152, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Crea, F.; Clermont, P.L.; Parolia, A.; Wang, Y.; Helgason, C.D. The non-coding transcriptome as a dynamic regulator of cancer metastasis. Cancer Metastasis Rev. 2014, 33, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Malone, C.D.; Hannon, G.J. Small RNAs as guardians of the genome. Cell 2009, 136, 656–668. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.S.; Zhou, R.; Rana, T.M. Gene regulation by non-coding RNAs. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.D.; Naldini, L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat. Rev. Genet. 2009, 10, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Bioinform. MicroRNA Res. 2017, 1617, 57–67. [Google Scholar]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Huang, J.; Yang, N.; Greshock, J.; Megraw, M.S.; Giannakakis, A.; Liang, S.; Naylor, T.L.; Barchetti, A.; Ward, M.R.; et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. USA 2006, 103, 9136–9141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, B.; Stresemann, C.; Brueckner, B.; Lyko, F. Methylation of Human MicroRNA Genes in Normal and Neoplastic Cells. Cell Cycle 2007, 6, 1001–1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Hsu, S.; Wang, X.; Kutay, H.; Bid, H.K.; Yu, J.; Ganju, R.K.; Jacob, S.T.; Yuneva, M.; Ghoshal, K. Reciprocal regulation of microRNA-122 and c-Myc in hepatocellular cancer: Role of E2F1 and transcription factor dimerization partner 2. Hepatology 2014, 59, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Donzelli, S.; Mori, F.; Bellissimo, T.; Sacconi, A.; Casini, B.; Frixa, T.; Roscilli, G.; Aurisicchio, L.; Facciolo, F.; Pompili, A.; et al. Epigenetic silencing of miR-145-5p contributes to brain metastasis. Oncotarget 2015, 6, 35183–35201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walz, A.L.; Ooms, A.; Gadd, S.; Gerhard, D.S.; Smith, M.A.; Guidry Auvil, J.M.; Meerzaman, D.; Chen, Q.R.; Hsu, C.H.; Yan, C.; et al. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell 2015, 27, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Fu, H.; Tie, Y.; Hu, Z.; Kong, W.; Wu, Y.; Zheng, X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009, 275, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.S.; Pressman, S.; Andress, A.P.; Kim, K.; White, J.L.; Cassidy, J.J.; Li, X.; Lubell, K.; Lim, D.H.; Cho, I.S.; et al. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 2009, 11, 1150–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stalder, L.; Heusermann, W.; Sokol, L.; Trojer, D.; Wirz, J.; Hean, J.; Fritzsche, A.; Aeschimann, F.; Pfanzagl, V.; Basselet, P.; et al. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J. 2013, 32, 1115–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barman, B.; Bhattacharyya, S.N. mRNA Targeting to Endoplasmic Reticulum Precedes Ago Protein Interaction and MicroRNA (miRNA)-mediated Translation Repression in Mammalian Cells. J. Biol. Chem. 2015, 290, 24650–24656. [Google Scholar] [CrossRef] [PubMed]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Janas, T.; Janas, M.M.; Sapoń, K.; Janas, T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015, 589, 1391–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tewari, M. A functional extracellular transcriptome in animals? Implications for biology, disease and medicine. Genome Biol. 2015, 16, 47. [Google Scholar] [CrossRef] [PubMed]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef] [PubMed]
- Neviani, P.; Fabbri, M. Exosomic microRNAs in the Tumor Microenvironment. Front. Med. 2015, 2, 47. [Google Scholar] [CrossRef] [PubMed]
- Caruso Bavisotto, C.; Marino Gammazza, A.; Rappa, F.; Fucarino, A.; Pitruzzella, A.; David, S.; Campanella, C. Exosomes: Can doctors still ignore their existence? EuroMediterr. Biomed. J. 2013, 8. [Google Scholar] [CrossRef]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta-Gen. Subj. 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Blostein, R.; Johnstone, R.M. Loss of the transferrin receptor during the maturation of sheep reticulocytes in vitro. An immunological approach. Biochem. J. 1983, 210, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Logozzi, M.; De Milito, A.; Lugini, L.; Borghi, M.; Calabrò, L.; Spada, M.; Perdicchio, M.; Marino, M.L.; Federici, C.; Iessi, E.; et al. High Levels of Exosomes Expressing CD63 and Caveolin-1 in Plasma of Melanoma Patients. PLoS ONE 2009, 4, e5219. [Google Scholar] [CrossRef] [PubMed]
- Cappello, F.; Logozzi, M.; Campanella, C.; Caruso Bavisotto, C.; Marcilla, A.; Properzi, F.; Fais, S. Exosome levels in human body fluids: A tumor marker by themselves? Eur. J. Pharm. Sci. 2017, 96, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A common pathway for a specialized function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhang, H.; Li, W.; Deng, Y.; Munegowda, M.A.; Chibbar, R.; Qureshi, M.; Xiang, J. Dendritic cells recruit T cell exosomes via exosomal LFA-1 leading to inhibition of CD8+ CTL responses through downregulation of peptide/MHC class I and Fas ligand-mediated cytotoxicity. J. Immunol. 2010, 185, 5268–5278. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Bhatnagar, S. Exosome function: From tumor immunology to pathogen biology. Traffic 2008, 9, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.-S.; Lee, J.-M.; Park, G.W.; Lim, H.-W.; Bang, J.Y.; Kim, Y.-K.; Kwon, K.-H.; Kwon, H.J.; Kim, K.P.; Gho, Y.S. Proteomic analysis of microvesicles derived from human colorectal cancer cells. J. Proteome Res. 2007, 6, 4646–4655. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Franchi, L.; Nunez, G.; Dubyak, G.R. Nonclassical IL-1 β secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J. Immunol. 2007, 179, 1913–1925. [Google Scholar] [CrossRef] [PubMed]
- Phoonsawat, W.; Aoki-Yoshida, A.; Tsuruta, T.; Sonoyama, K. Adiponectin is partially associated with exosomes in mouse serum. Biochem. Biophys. Res. Commun. 2014, 448, 261–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Chen, R.; Kemper, S.; Charrier, A.; Brigstock, D.R. Suppression of fibrogenic signaling in hepatic stellate cells by Twist1-dependent microRNA-214 expression: Role of exosomes in horizontal transfer of Twist1. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G491–G499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campanella, C.; Bucchieri, F.; Merendino, A.M.; Fucarino, A.; Burgio, G.; Corona, D.F.V.; Barbieri, G.; David, S.; Farina, F.; Zummo, G.; et al. The odyssey of Hsp60 from tumor cells to other destinations includes plasma membrane-associated stages and Golgi and exosomal protein-trafficking modalities. PLoS ONE 2012, 7, e42008. [Google Scholar] [CrossRef] [PubMed]
- Campanella, C.; Rappa, F.; Sciumè, C.; Marino Gammazza, A.; Barone, R.; Bucchieri, F.; David, S.; Curcurù, G.; Caruso Bavisotto, C.; Pitruzzella, A.; et al. Heat shock protein 60 levels in tissue and circulating exosomes in human large bowel cancer before and after ablative surgery. Cancer 2015. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, G.I.; Febbraio, M.A. Exosome-dependent trafficking of Hsp70: A novel secretory pathway for cellular stress proteins. J. Biol. Chem. 2005, 280, 23349–23355. [Google Scholar] [CrossRef] [PubMed]
- Bausero, M.A.; Gastpar, R.; Multhoff, G.; Asea, A. Alternative mechanism by which IFN-gamma enhances tumor recognition: Active release of heat shock protein 72. J. Immunol. 2005, 175, 2900–2912. [Google Scholar] [CrossRef] [PubMed]
- Vega, V.L.; Rodríguez-Silva, M.; Frey, T.; Gehrmann, M.; Diaz, J.C.; Steinem, C.; Multhoff, G.; Arispe, N.; De Maio, A. Hsp70 translocates into the plasma membrane after stress and is released into the extracellular environment in a membrane-associated form that activates macrophages. J. Immunol. 2008, 180, 4299–4307. [Google Scholar] [CrossRef] [PubMed]
- Gastpar, R.; Gehrmann, M.; Bausero, M.A.; Asea, A.; Gross, C.; Schroeder, J.A.; Multhoff, G. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005, 65, 5238–5247. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.-H.; Wan, Y.-L.; Lin, Y.; Zhang, W.; Yang, M.; Li, G.-L.; Lin, H.-M.; Shang, C.-Z.; Chen, Y.-J.; Min, J. Anticancer drugs cause release of exosomes with heat shock proteins from human hepatocellular carcinoma cells that elicit effective natural killer cell antitumor responses in vitro. J. Biol. Chem. 2012, 287, 15874–15885. [Google Scholar] [CrossRef] [PubMed]
- McCready, J.; Sims, J.D.; Chan, D.; Jay, D.G. Secretion of extracellular Hsp90α via exosomes increases cancer cell motility: A role for plasminogen activation. BMC Cancer 2010, 10, 294. [Google Scholar] [CrossRef] [PubMed]
- Merendino, A.M.; Bucchieri, F.; Campanella, C.; Marcianò, V.; Ribbene, A.; David, S.; Zummo, G.; Burgio, G.; Corona, D.F.V.; Conway de Macario, E.; et al. Hsp60 is actively secreted by human tumor cells. PLoS ONE 2010, 5, e9247. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Knowlton, A.-A. Hsp60 trafficking in adult cardiac myocytes: Role of the exosomal pathway. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H3052–H3056. [Google Scholar] [CrossRef] [PubMed]
- Hayoun, D.; Kapp, T.; Edri-Brami, M.; Ventura, T.; Cohen, M.; Avidan, A.; Lichtenstein, R.G. Hsp60 is transported through the secretory pathway of 3-MCA-induced fibrosarcoma tumour cells and undergoes N-glycosylation. FEBS J. 2012, 279, 2083–2095. [Google Scholar] [CrossRef] [PubMed]
- Marino Gammazza, A.; Campanella, C.; Barone, R.; Caruso Bavisotto, C.; Gorska, M.; Wozniak, M.; Carini, F.; Cappello, F.; D’Anneo, A.; Lauricella, M.; et al. Doxorubicin anti-tumor mechanisms include Hsp60 post-translational modifications leading to the Hsp60/p53 complex dissociation and instauration of replicative senescence. Cancer Lett. 2017, 385, 75–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Guduric-Fuchs, J.; O’Connor, A.; Camp, B.; O’Neill, C.L.; Medina, R.J.; Simpson, D.A. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012, 13, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nolte’T Hoen, E.N.M.; Buermans, H.P.J.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.M.; ’T Hoen, P.A.C. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.C.Y.; Eaton, S.A.; Young, P.E.; Lee, M.; Shuttleworth, R.; Humphreys, D.T.; Grau, G.E.; Combes, V.; Bebawy, M.; Gong, J.; et al. Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol. 2013, 10, 1333–1344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiera, G.; Di Liegro, C.M.; Di Liegro, I. Molecular Determinants of Malignant Brain Cancers: From Intracellular Alterations to Invasion Mediated by Extracellular Vesicles. Int. J. Mol. Sci. 2017, 18, 2774. [Google Scholar] [CrossRef] [PubMed]
- Ngolab, J.; Trinh, I.; Rockenstein, E.; Mante, M.; Florio, J.; Trejo, M.; Masliah, D.; Adame, A.; Masliah, E.; Rissman, R.A. Brain-derived exosomes from dementia with Lewy bodies propagate α-synuclein pathology. Acta Neuropathol. Commun. 2017, 5, 46. [Google Scholar] [CrossRef] [PubMed]
- Picciolini, S.; Gualerzi, A.; Vanna, R.; Sguassero, A.; Gramatica, F.; Bedoni, M.; Masserini, M.; Morasso, C. Detection and Characterization of Different Brain-Derived Subpopulations of Plasma Exosomes by Surface Plasmon Resonance Imaging. Anal. Chem. 2018, 90, 8873–8880. [Google Scholar] [CrossRef] [PubMed]
- Karnati, H.K.; Garcia, J.H.; Tweedie, D.; Becker, R.E.; Kapogiannis, D.; Greig, N.H. Neuronal Enriched Extracellular Vesicle Proteins as Biomarkers for Brain Traumatic Injury. J. Neurotrauma 2018. [Google Scholar] [CrossRef] [PubMed]
- Mrowczynski, O.D.; Zacharia, B.E.; Connor, J.R. Exosomes and their implications in central nervous system tumor biology. Prog. Neurobiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Liu, L.; Ma, F.; Wong, C.W.; Guo, X.E.; Chacko, J.V.; Farhoodi, H.P.; Zhang, S.X.; Zimak, J.; Ségaliny, A.; et al. Elucidation of Exosome Migration Across the Blood–Brain Barrier Model In Vitro. Cell. Mol. Bioeng. 2016, 9, 509–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, D.; Zhao, Y.; Banks, W.A.; Bullock, K.M.; Haney, M.; Batrakova, E.; Kabanov, A.V. Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain. Biomaterials 2017, 142, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Graner, M.; Redzic, J.; Ung, T. Glioblastoma extracellular vesicles: Reservoirs of potential biomarkers. Pharmgenom. Pers. Med. 2014, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.; Ahn, S.; Yeom, C.-H.; Park, S. Exosome Proteome of U-87MG Glioblastoma Cells. Biology 2016, 5, 50. [Google Scholar] [CrossRef] [PubMed]
- Mallawaaratchy, D.M.; Hallal, S.; Russell, B.; Ly, L.; Ebrahimkhani, S.; Wei, H.; Christopherson, R.I.; Buckland, M.E.; Kaufman, K.L. Comprehensive proteome profiling of glioblastoma-derived extracellular vesicles identifies markers for more aggressive disease. J. Neurooncol. 2017, 131, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W. T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimoto, K.; Dang, J.; Zhu, S.; Nathanson, D.; Huang, T.; Dumont, R.; Seligson, D.B.; Yong, W.H.; Xiong, Z.; Rao, N.; et al. Development of a real-time RT-PCR assay for detecting EGFRvIII in glioblastoma samples. Clin. Cancer Res. 2008, 14, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Strik, H.M.; Weller, M.; Frank, B.; Hermisson, M.; Deininger, M.H.; Dichgans, J.; Meyermann, R. Heat shock protein expression in human gliomas. Anticancer Res. 2000, 20, 4457–4462. [Google Scholar] [PubMed]
- Shen, G.; Liang, S.; Xu, Z.; Zhou, L.; Xiao, S.; Xia, X.; Li, R.; Liao, Y.; You, C.; Wei, Y. Downregulated expression of Hsp27 in human low-grade glioma tissues discovered by a quantitative proteomic analysis. Proteome Sci. 2010, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.A.; Kabapy, N.F.; Deraz, S.F.; Smith, C. Heat shock proteins in oncology: Diagnostic biomarkers or therapeutic targets? Biochim. Biophys. Acta 2011, 1816, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Golembieski, W.A.; Thomas, S.L.; Schultz, C.R.; Yunker, C.K.; McClung, H.M.; Lemke, N.; Cazacu, S.; Barker, T.; Sage, E.H.; Brodie, C.; et al. Hsp27 mediates SPARC-induced changes in glioma morphology, migration, and invasion. Glia 2008, 56, 1061–1075. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Xu, Y.; Guan, D.; Liu, Z.; Liu, D.X. Hsp70 protein promotes survival of C6 and U87 glioma cells by inhibition of ATF5 degradation. J. Biol. Chem. 2011, 286, 20251–20259. [Google Scholar] [CrossRef] [PubMed]
- Sauvageot, C.M.-E.; Weatherbee, J.L.; Kesari, S.; Winters, S.E.; Barnes, J.; Dellagatta, J.; Ramakrishna, N.R.; Stiles, C.D.; Kung, A.L.-J.; Kieran, M.W.; et al. Efficacy of the Hsp90 inhibitor 17-AAG in human glioma cell lines and tumorigenic glioma stem cells. Neuro. Oncol. 2009, 11, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Nagaraju, G.P.; Long, T.-E.; Park, W.; Landry, J.C.; Taliaferro-Smith, L.; Farris, A.B.; Diaz, R.; El-Rayes, B.F. Heat shock protein 90 promotes epithelial to mesenchymal transition, invasion, and migration in colorectal cancer. Mol. Carcinog. 2015, 54, 1147–1158. [Google Scholar] [CrossRef] [PubMed]
- Rappa, F.; Cappello, F.; Halatsch, M.-E.; Scheuerle, A.; Kast, R.E. Aldehyde dehydrogenase and Hsp90 co-localize in human glioblastoma biopsy cells. Biochimie 2013, 95, 782–786. [Google Scholar] [CrossRef] [PubMed]
- Rappa, F.; Unti, E.; Baiamonte, P.; Cappello, F.; Scibetta, N. Different immunohistochemical levels of Hsp60 and Hsp70 in a subset of brain tumors and putative role of Hsp60 in neuroepithelial tumorigenesis. Eur. J. Histochem. 2013, 57, e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clayton, A.; Turkes, A.; Navabi, H.; Mason, M.D.; Tabi, Z. Induction of heat shock proteins in B-cell exosomes. J. Cell Sci. 2005, 118, 3631–3638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campanella, C.; Caruso Bavisotto, C.; Marino Gammazza, A.; Nikolic, D.; Rappa, F.; David, S.; Cappello, F.; Bucchieri, F.; Fais, S. Exosomal Heat Shock Proteins as New Players in Tumour Cell-to-cell Communication. J. Circ. Biomark. 2014, 3, 4. [Google Scholar] [CrossRef] [Green Version]
- Gimenez, M.; Marie, S.K.N.; Oba-Shinjo, S.; Uno, M.; Izumi, C.; Oliveira, J.B.; Rosa, J.C. Quantitative proteomic analysis shows differentially expressed HspB1 in glioblastoma as a discriminating short from long survival factor and NOVA1 as a differentiation factor between low-grade astrocytoma and oligodendroglioma. BMC Cancer 2015, 15, 481. [Google Scholar] [CrossRef] [PubMed]
- Thuringer, D.; Hammann, A.; Benikhlef, N.; Fourmaux, E.; Bouchot, A.; Wettstein, G.; Solary, E.; Garrido, C. Transactivation of the epidermal growth factor receptor by heat shock protein 90 via Toll-like receptor 4 contributes to the migration of glioblastoma cells. J. Biol. Chem. 2011, 286, 3418–3828. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Li, Y.; Hou, X.; Zhang, N.; Ma, J.; Ding, F.; Li, F.; Miao, Z.; Zhang, Y.; Qi, Q.; et al. Hsp60 is involved in the neuroprotective effects of naloxone. Mol. Med. Rep. 2014, 10, 2172–2176. [Google Scholar] [CrossRef] [PubMed]
- Mäkelä, K.S.; Haapasalo, J.A.; Ilvesaro, J.M.; Parkkila, S.; Paavonen, T.; Haapasalo, H.K. Hsp27 and its expression pattern in diffusely infiltrating astrocytomas. Histol. Histopathol. 2014, 29, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Graner, M.W.; Raynes, D.A.; Bigner, D.D.; Guerriero, V. Heat shock protein 70-binding protein 1 is highly expressed in high-grade gliomas, interacts with multiple heat shock protein 70 family members, and specifically binds brain tumor cell surfaces. Cancer Sci. 2009, 100, 1870–1879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morino, M.; Tsuzuki, T.; Ishikawa, Y.; Shirakami, T.; Yoshimura, M.; Kiyosuke, Y.; Matsunaga, K.; Yoshikumi, C.; Saijo, N. Specific expression of Hsp27 in human tumor cell lines in vitro. In Vivo 1997, 11, 179–184. [Google Scholar] [PubMed]
- Scott, K.A.; Dennis, J.L.; Dalgleish, A.G.; Liu, W.M. Inhibiting Heat Shock Proteins Can Potentiate the Cytotoxic Effect of Cannabidiol in Human Glioma Cells. Anticancer Res. 2015, 35, 5827–5837. [Google Scholar] [PubMed]
- Ghosh, J.C.; Siegelin, M.D.; Dohi, T.; Altieri, D.C. Heat shock protein 60 regulation of the mitochondrial permeability transition pore in tumor cells. Cancer Res. 2010, 70, 8988–8993. [Google Scholar] [CrossRef] [PubMed]
- Alexiou, G.A.; Vartholomatos, G.; Stefanaki, K.; Patereli, A.; Dova, L.; Karamoutsios, A.; Lallas, G.; Sfakianos, G.; Moschovi, M.; Prodromou, N. Expression of heat shock proteins in medulloblastoma. J. Neurosurg. Pediatr. 2013, 12, 452–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marino Gammazza, A.; Caruso Bavisotto, C.; David, S.; Barone, R.; Rappa, F.; Campanella, C.; Conway de Macario, E.; Cappello, F.; Macario, A.J.L. Hsp60 is a ubiquitous player in the physiological and pathogenic interactions between the chaperoning and the immune systems. Curr. Immunol. Rev. 2017, 13, 44–45. [Google Scholar] [CrossRef]
- Samali, A.; Cai, J.; Zhivotovsky, B.; Jones, D.P.; Orrenius, S. Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of jurkat cells. EMBO J. 1999, 18, 2040–2048. [Google Scholar] [CrossRef] [PubMed]
- Caruso Bavisotto, C.; Nikolic, D.; Marino Gammazza, A.; Barone, R.; Lo Cascio, F.; Mocciaro, E.; Zummo, G.; Conway de Macario, E.; Macario, A.J.L.; Cappello, F.; et al. The dissociation of the Hsp60/pro-Caspase-3 complex by bis(pyridyl)oxadiazole copper complex (CubipyOXA) leads to cell death in NCI-H292 cancer cells. J. Inorg. Biochem. 2017, 170, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, S.R.; Gupta, S.; Knowlton, A.A. Cytosolic heat shock protein 60, apoptosis, and myocardial injury. Circulation 2002, 105, 2899–2904. [Google Scholar] [CrossRef] [PubMed]
- Stefano, L.; Racchetti, G.; Bianco, F.; Passini, N.; Gupta, R.S.; Panina Bordignon, P.; Meldolesi, J. The surface-exposed chaperone, Hsp60, is an agonist of the microglial TREM2 receptor. J. Neurochem. 2009, 110, 284–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Creagh, E.M.; Carmody, R.J.; Cotter, T.G. Heat shock protein 70 inhibits caspase-dependent and -independent apoptosis in Jurkat T cells. Exp. Cell Res. 2000, 257, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Belkacemi, L.; Hebb, M.O. Hsp27 knockdown produces synergistic induction of apoptosis by Hsp90 and kinase inhibitors in glioblastoma multiforme. Anticancer Res. 2014, 34, 4915–4927. [Google Scholar] [PubMed]
- Wu, Z.B.; Cai, L.; Lin, S.J.; Leng, Z.G.; Guo, Y.H.; Yang, W.L.; Chu, Y.W.; Yang, S.-H.; Zhao, W.G. Heat Shock Protein 47 Promotes Glioma Angiogenesis. Brain Pathol. 2016, 26, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Trentin, G.A.; He, Y.; Wu, D.C.; Tang, D.; Rozakis-Adcock, M. Identification of a hTid-1 mutation which sensitizes gliomas to apoptosis. FEBS Lett. 2004, 578, 323–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, H.; Komatsuda, A.; Wakui, H.; Miura, A.B.; Tashima, Y. Mammalian Hsp60 is a major target for an immunosuppressant mizoribine. J. Biol. Chem. 1999, 274, 35147–35151. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, R.; Sugihara, T.; Yoshida, A.; Nomura, H.; Reddel, R.R.; Simpson, R.; Maruta, H.; Kaul, S.C. Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res. 2000, 60, 6818–6821. [Google Scholar] [PubMed]
- Jego, G.; Hazoumé, A.; Seigneuric, R.; Garrido, C. Targeting heat shock proteins in cancer. Cancer Lett. 2013, 332, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Lamberti, D.; Cristinziano, G.; Porru, M.; Leonetti, C.; Egan, J.B.; Shi, C.-X.; Buglioni, S.; Amoreo, C.A.; Castellani, L.; Borad, M.J.; et al. Hsp90 inhibition drives degradation of FGFR2 fusion proteins: Implications for treatment of cholangiocarcinoma. Hepatology 2018. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Jiang, F.; Zhou, J.; Wu, D.; Sheng, Z.; Li, M. Hsp90: A Novel Target Gene of miRNA-628-3p in A549 Cells. Biomed Res. Int. 2018, 2018, 4149707. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Cai, M.; Fu, D.; Chen, K.; Sun, M.; Cai, Z.; Cheng, B. Heat shock protein 90B1 plays an oncogenic role and is a target of microRNA-223 in human osteosarcoma. Cell. Physiol. Biochem. 2012, 30, 1481–1490. [Google Scholar] [CrossRef] [PubMed]
- Kariya, A.; Furusawa, Y.; Yunoki, T.; Kondo, T.; Tabuchi, Y. A microRNA-27a mimic sensitizes human oral squamous cell carcinoma HSC-4 cells to hyperthermia through downregulation of Hsp110 and Hsp90. Int. J. Mol. Med. 2014, 34, 334–340. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, T.N.; Mujumdar, N.; Banerjee, S.; Sangwan, V.; Sarver, A.; Vickers, S.; Subramanian, S.; Saluja, A.K. Triptolide induces the expression of miR-142-3p: A negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Mol. Cancer Ther. 2013, 12, 1266–1275. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.-X.; Lin, Q.-X.; Deng, C.-Y.; Zhu, J.-N.; Mai, L.-P.; Liu, J.-L.; Fu, Y.-H.; Liu, X.-Y.; Li, Y.-X.; Zhang, Y.-Y.; et al. miR-1/miR-206 regulate Hsp60 expression contributing to glucose-mediated apoptosis in cardiomyocytes. FEBS Lett. 2010, 584, 3592–3600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Xie, T.; Xue, N.; Kuang, Q.; Wei, Z.; Liang, M.; Ding, X. miR-382 Contributes to Renal Tubulointerstitial Fibrosis by Downregulating HSPD1. Oxid. Med. Cell. Longev. 2017, 2017, 4708516. [Google Scholar] [CrossRef] [PubMed]
- Choghaei, E.; Khamisipour, G.; Falahati, M.; Naeimi, B.; Mossahebi-Mohammadi, M.; Tahmasebi, R.; Hasanpour, M.; Shamsian, S.; Hashemi, Z.S. Knockdown of microRNA-29a Changes the Expression of Heat Shock Proteins in Breast Carcinoma MCF-7 Cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2016, 23, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, G.M.V; Paratore, S.; Caltabiano, R.; Palmucci, S.; Parra, H.S.; Privitera, G.; Motta, F.; Lanzafame, S.; Scaglione, G.; Longo, A.; et al. Long-term therapy with temozolomide is a feasible option for newly diagnosed glioblastoma: A single-institution experience with as many as 101 temozolomide cycles. Neurosurg. Focus 2014, 37, E4. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, R.; Villa, A.; Pino, M.; Imperato, A.; Giammalva, G.R.; Costantino, G.; Graziano, F.; Gulì, C.; Meli, F.; Francaviglia, N.; et al. With a little help from my friends: The role of intraoperative fluorescent dyes in the surgical management of high-grade gliomas. Brain Sci. 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, S.; Wangtao; Wang, Y.; Wang, J.; Jiang, L.; Li, S.; Hu, X.; Wang, Q. Upregulation of GRP78 and GRP94 and its function in chemotherapy resistance to VP-16 in human lung cancer cell line SK-MES-1. Cancer Investig. 2009, 27, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Wang, S.; Wu, D.; Yang, W. Decreased functional expression of Grp78 and Grp94 inhibits proliferation and attenuates apoptosis in a human gastric cancer cell line in vitro. Oncol. Lett. 2015, 9, 1181–1186. [Google Scholar] [CrossRef] [PubMed]
- Masui, K.; Cloughesy, T.F.; Mischel, P.S. Molecular pathology in adult high-grade gliomas: From molecular diagnostics to target therapies. Neuropathol. Appl. Neurobiol. 2012, 38, 271–291. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Lu, Y.; Pan, Z.; Chu, W.; Luo, X.; Lin, H.; Xiao, J.; Shan, H.; Wang, Z.; Yang, B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting Hsp60, Hsp70 and caspase-9 in cardiomyocytes. J. Cell Sci. 2007, 120, 3045–3052. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Sun, X.; Ren, J.; Li, X.; Gao, X.; Lu, C.; Zhang, Y.; Sun, H.; Wang, Y.; Wang, H.; et al. miR-1 Exacerbates Cardiac Ischemia-Reperfusion Injury in Mouse Models. PLoS ONE 2012, 7, e50515. [Google Scholar] [CrossRef] [PubMed]
- Neumann, E.; Brandenburger, T.; Santana-Varela, S.; Deenen, R.; Köhrer, K.; Bauer, I.; Hermanns, H.; Wood, J.N.; Zhao, J.; Werdehausen, R. MicroRNA-1-associated effects of neuron-specific brain-derived neurotrophic factor gene deletion in dorsal root ganglia. Mol. Cell. Neurosci. 2016, 75, 36–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evert, B.O.; Nalavade, R.; Jungverdorben, J.; Matthes, F.; Weber, S.; Rajput, A.; Bonn, S.; Brüstle, O.; Peitz, M.; Krauß, S. Upregulation of miR-370 and miR-543 is associated with reduced expression of heat shock protein 40 in spinocerebellar ataxia type 3. PLoS ONE 2018, 13, e0201794. [Google Scholar] [CrossRef] [PubMed]
- Graziano, F.; Caruso Bavisotto, C.; Marino Gammazza, A.; Rappa, F.; Conway de Macario, E.; Macario, A.J.L.; Cappello, F.; Campanella, C.; Maugeri, R.; Iacopino, D.G. Chaperonology: The third eye on brain gliomas. Brain Sci. 2018, 8. [Google Scholar] [CrossRef] [PubMed]

| Main GBM-Related Chaperones 1 | Functions | Ref. |
|---|---|---|
| Hsp27 | Phosphorylated Hsp27 at high levels co-localizes with secreted protein acidic and rich in cysteine (SPARC), and associates with changes in cell morphology, migration, and invasion in vitro. | [102] |
| Hsp27 overexpression is observed in parallel to the increase in malignancy, as a predictive factor of poor prognosis for GBM. | [111] | |
| Hsp60 | Hsp60 overexpression inhibit tumor cell death or antitumor immune system response. | [108] |
| In glioma cell line, Hsp60 has an antiapoptotic function through CypD-mediated mitochondrial permeability regulation. | [112] | |
| Hsp60 binds triggering receptor expressed in myeloid cells 2 (TREM2), increasing phagocytic activity in the N9 microglial cell line. | [113] | |
| Hsp70 | Hsp70 stabilize the activating transcription factor 5 (ATF5), determining a pro-survival effect in C6 and U87 cells. | [104] |
| Hsp70 overexpression inhibits tumor cell death or antitumor immune system response. | [108] | |
| Hsp90 | Hsp90 binds and stabilize its client protein (e.g., PTEN, p53, and EGFR) to maintain its expression, leading to aggressive growth in GBM. | [105] |
| Hsp90 co-localize with ALDH (aldehyde dehydrogenase) in cancer stem cell sub-population. | [107] | |
| Extracellular Hsp90α favors cell migration of glioblastoma U87 cells. | [112] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caruso Bavisotto, C.; Graziano, F.; Rappa, F.; Marino Gammazza, A.; Logozzi, M.; Fais, S.; Maugeri, R.; Bucchieri, F.; Conway de Macario, E.; Macario, A.J.L.; et al. Exosomal Chaperones and miRNAs in Gliomagenesis: State-of-Art and Theranostics Perspectives. Int. J. Mol. Sci. 2018, 19, 2626. https://doi.org/10.3390/ijms19092626
Caruso Bavisotto C, Graziano F, Rappa F, Marino Gammazza A, Logozzi M, Fais S, Maugeri R, Bucchieri F, Conway de Macario E, Macario AJL, et al. Exosomal Chaperones and miRNAs in Gliomagenesis: State-of-Art and Theranostics Perspectives. International Journal of Molecular Sciences. 2018; 19(9):2626. https://doi.org/10.3390/ijms19092626
Chicago/Turabian StyleCaruso Bavisotto, Celeste, Francesca Graziano, Francesca Rappa, Antonella Marino Gammazza, Mariantonia Logozzi, Stefano Fais, Rosario Maugeri, Fabio Bucchieri, Everly Conway de Macario, Alberto J. L. Macario, and et al. 2018. "Exosomal Chaperones and miRNAs in Gliomagenesis: State-of-Art and Theranostics Perspectives" International Journal of Molecular Sciences 19, no. 9: 2626. https://doi.org/10.3390/ijms19092626
APA StyleCaruso Bavisotto, C., Graziano, F., Rappa, F., Marino Gammazza, A., Logozzi, M., Fais, S., Maugeri, R., Bucchieri, F., Conway de Macario, E., Macario, A. J. L., Cappello, F., Iacopino, D. G., & Campanella, C. (2018). Exosomal Chaperones and miRNAs in Gliomagenesis: State-of-Art and Theranostics Perspectives. International Journal of Molecular Sciences, 19(9), 2626. https://doi.org/10.3390/ijms19092626