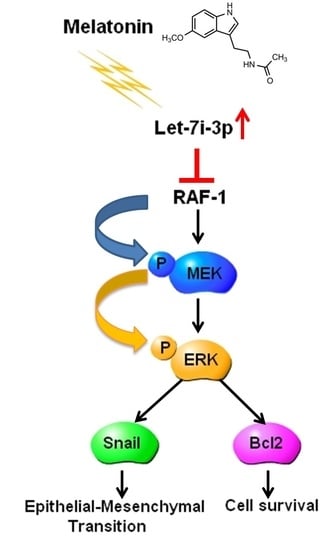
Melatonin Inhibits the Progression of Hepatocellular Carcinoma through MicroRNA Let7i-3p Mediated RAF1 Reduction
Abstract
:1. Introduction
2. Results
2.1. Melatonin Inhibits HCC-Cell Proliferation, Migration and Invasiveness
2.2. Melatonin Induces miRNA Let7i-3p Expression
2.3. Let7i-3p Acts as a Tumor Suppressor
2.4. Let7i-3p Directly Binds to RAF1 mRNA 3′UTR and Suppresses RAF1 Expression
2.5. Melatonin Reduces RAF1 Expression and Its Downstream Oncogenic MAPK Signaling
2.6. Melatonin Inhibits HCC Progression through Let7i-3p-Mediated RAF1 Suppression
2.7. Melatonin Suppresses Tumor Growth In Vivo
3. Discussion
4. Material and Methods
4.1. Human Specimens
4.2. Cell Lines, Antibodies, miRNAs and Plasmids
4.3. Cell Proliferation Assay
4.4. Cell Migration and Invasion Assays
4.5. RNA Isolation
4.6. Library Preparation and Sequencing
4.7. Small-RNA Sequencing Analysis
4.8. Detection of MiRNA Let7i-3p through Quantitative Real-Time RT-PCR
4.9. Estimation of Protein Levels through Western Blotting
4.10. Transient Transfection of the pmirGLO-RAF1 Plasmid and miRNAs
4.11. Mice
4.12. In Vivo Tumor Growth Assay
4.13. Immunohistochemistry
4.14. Data Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McGlynn, K.A.; Petrick, J.L.; London, W.T. Global epidemiology of hepatocellular carcinoma: An emphasis on demographic and regional variability. Clin. Liver Dis. 2015, 19, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Ghouri, Y.A.; Mian, I.; Rowe, J.H. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J. Carcinog. 2017, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Wörns, M.A.; Galle, P.R. HCC therapies—Lessons learned. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Sood, G.K. Hepatocellular carcinoma review: Current treatment, and evidence-based medicine. World J. Gastroenterol. 2014, 20, 4115–4127. [Google Scholar] [CrossRef] [PubMed]
- Lochner, A.; Marais, E.; Huisamen, B. Melatonin and cardioprotection against ischaemia/reperfusion injury: What’s new? A review. J. Pineal Res. 2018, 65, e12490. [Google Scholar] [CrossRef] [PubMed]
- Mortezaee, K.; Khanlarkhani, N. Melatonin application in targeting oxidative-induced liver injuries: A review. J. Cell. Physiol. 2018, 233, 4015–4032. [Google Scholar] [CrossRef] [PubMed]
- Favero, G.; Franceschetti, L.; Bonomini, F.; Rodella, L.F.; Rezzani, R. Melatonin as an anti-inflammatory agent modulating inflammasome activation. Int. J. Endocrinol. 2017, 2017, 1835195. [Google Scholar] [CrossRef] [PubMed]
- Osier, N.; McGreevy, E.; Pham, L.; Puccio, A.; Ren, D.; Conley, Y.P.; Alexander, S.; Dixon, C.E. Melatonin as a therapy for traumatic brain injury: A review of published evidence. Int. J. Mol. Sci. 2018, 19, 1539. [Google Scholar] [CrossRef] [PubMed]
- Su, S.C.; Hsieh, M.J.; Yang, W.E.; Chung, W.H.; Reiter, R.J.; Yang, S.F. Cancer metastasis: Mechanisms of inhibition by melatonin. J. Pineal Res. 2017, 62, e12370. [Google Scholar] [CrossRef] [PubMed]
- Acuña-Castroviejo, D.; Escames, G.; Venegas, C.; Díaz-Casado, M.E.; Lima-Cabello, E.; López, L.C.; Rosales-Corral, S.; Tan, D.X.; Reiter, R.J. Extrapineal melatonin: Sources, regulation, and potential functions. Cell. Mol. Life Sci. 2014, 71, 2997–3025. [Google Scholar] [CrossRef] [PubMed]
- Bubenik, G.A. Thirty four years since the discovery of gastrointestinal melatonin. J. Physiol. Pharmacol. 2008, 59 (Suppl. 2), 33–51. [Google Scholar]
- Dominguez-Rodriguez, A.; Abreu-Gonzalez, P.; Sanchez-Sanchez, J.J.; Kaski, J.C.; Reiter, R.J. Melatonin and circadian biology in human cardiovascular disease. J. Pineal Res. 2010, 49, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Vico, A.; Lardone, P.J.; Alvarez-Sánchez, N.; Rodríguez-Rodríguez, A.; Guerrero, J.M. Melatonin: Buffering the immune system. Int. J. Mol. Sci. 2013, 14, 8638–8683. [Google Scholar] [CrossRef] [PubMed]
- Jaworek, J.; Leja-Szpak, A.; Nawrot-Porąbka, K.; Szklarczyk, J.; Kot, M.; Pierzchalski, P.; Góralska, M.; Ceranowicz, P.; Warzecha, Z.; Dembinski, A.; et al. Effects of melatonin and its analogues on pancreatic inflammation, enzyme secretion, and tumorigenesis. Int. J. Mol. Sci. 2017, 18, 1014. [Google Scholar] [CrossRef] [PubMed]
- García, J.J.; López-Pingarrón, L.; Almeida-Souza, P.; Tres, A.; Escudero, P.; Garcia-Gil, F.A.; Tan, D.X.; Reiter, R.J.; Ramírez, J.M.; Bernal-Pérez, M. Protective effects of melatonin in reducing oxidative stress and in preserving the fluidity of biological membranes: A review. J. Pineal Res. 2014, 56, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Mayo, J.C.; Tan, D.X.; Sainz, R.M.; Alatorre-Jimenez, M.; Qin, L. Melatonin as an antioxidant: Under promises but over delivers. J. Pineal Res. 2016, 61, 253–278. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.X.; Manchester, L.C.; Hardeland, R.; Lopez-Burillo, S.; Mayo, J.C.; Sainz, R.M.; Reiter, R.J. Melatonin: A hormone, a tissue factor, an autocoid, a paracoid, and an antioxidant vitamin. J. Pineal Res. 2003, 34, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Nooshinfar, E.; Safaroghli-Azar, A.; Bashash, D.; Akbari, M.E. Melatonin, an inhibitory agent in breast cancer. Breast Cancer 2017, 24, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Borin, T.F.; Arbab, A.S.; Gelaleti, G.B.; Ferreira, L.C.; Moschetta, M.G.; Jardim-Perassi, B.V.; Iskander, A.S.; Varma, N.R.; Shankar, A.; Coimbra, V.B.; et al. Melatonin decreases breast cancer metastasis by modulating Rho-associated kinase protein-1 expression. J. Pineal Res. 2016, 60, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Yang, Y.; Fan, C.; Han, J.; Wang, D.; Di, S.; Hu, W.; Liu, D.; Li, X.; Reiter, R.J.; et al. Melatonin as a potential anticarcinogen for non-small-cell lung cancer. Oncotarget 2016, 7, 46768–46784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fic, M.; Gomulkiewicz, A.; Grzegrzolka, J.; Podhorska-Okolow, M.; Zabel, M.; Dziegiel, P.; Jablonska, K. The impact of melatonin on colon cancer cells’ resistance to doxorubicin in an in vitro study. Int. J. Mol. Sci. 2017, 18, 1396. [Google Scholar] [CrossRef] [PubMed]
- Cerea, G.; Vaghi, M.; Ardizzoia, A.; Villa, S.; Bucovec, R.; Mengo, S.; Gardani, G.; Tancini, G.; Lissoni, P. Biomodulation of cancer chemotherapy for metastatic colorectal cancer: A randomized study of weekly low-dose irinotecan alone versus irinotecan plus the oncostatic pineal hormone melatonin in metastatic colorectal cancer patients progressing on 5-fluorouracil-containing combinations. Anticancer Res. 2003, 23, 1951–1954. [Google Scholar] [PubMed]
- Bartel, D.P. Metazoan microRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Hirschberger, S.; Hinske, L.C.; Kreth, S. MiRNAs: Dynamic regulators of immune cell functions in inflammation and cancer. Cancer Lett. 2018, 431, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Nejad, C.; Stunden, H.J.; Gantier, M.P. A guide to miRNAs in inflammation and innate immune responses. FEBS J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Dykxhoorn, D.M. MicroRNAs and metastasis: Little RNAs go a long way. Cancer Res. 2010, 70, 6401–6406. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Mateescu, B.; Batista, L.; Cardon, M.; Gruosso, T.; de Feraudy, Y.; Mariani, O.; Nicolas, A.; Meyniel, J.P.; Cottu, P.; Sastre-Garau, X.; et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat. Med. 2011, 17, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Dagan, L.N.; Jiang, X.; Bhatt, S.; Cubedo, E.; Rajewsky, K.; Lossos, I.S. miR-155 regulates HGAL expression and increases lymphoma cell motility. Blood 2012, 119, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xue, J.L.; Shen, Q.; Chen, J.; Tian, L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 2012, 55, 1852–1862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Gregory, P.A.; Fernandes, R.C.; Denis, I.; Wang, Q.; Townley, S.L.; Zhao, S.G.; Hanson, A.R.; Pickering, M.A.; Armstrong, H.K.; et al. MicroRNA-194 promotes prostate cancer metastasis by inhibiting SOCS2. Cancer Res. 2017, 77, 1021–1034. [Google Scholar] [CrossRef] [PubMed]
- Smolle, M.A.; Leithner, A.; Posch, F.; Szkandera, J.; Liegl-Atzwanger, B.; Pichler, M. MicroRNAs in different histologies of soft tissue sarcoma: A comprehensive review. Int. J. Mol. Sci. 2017, 18, 1960. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Liu, Z.; Yang, X.; Zhou, J.; Yu, H.; Zhang, R.; Li, H. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018, 414, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, I.; Francis, H.; Meng, F.; Glaser, S.; Alpini, G. Diagnostic and therapeutic potentials of microRNAs in cholangiopathies. Liver Res. 2017, 1, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Duggal, B.; Gupta, M.K.; Naga Prasad, S.V. Potential role of microRNAs in cardiovascular disease: Are they up to their hype? Curr. Cardiol. Rev. 2016, 12, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.D.; Chang, S. Development of novel therapeutic agents by inhibition of oncogenic microRNAs. Int. J. Mol. Sci. 2017, 19, 65. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wang, J.; Liu, L.; Yu, L.; Zhao, N.; Zhou, X.; Lu, X. Curcumin increases the sensitivity of Paclitaxel-resistant NSCLC cells to Paclitaxel through microRNA-30c-mediated MTA1 reduction. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, C.; Peng, F.H.; Peng, L.K. MiR-200c sensitizes clear-cell renal cell carcinoma cells to sorafenib and imatinib by targeting heme oxygenase-1. Neoplasma 2014, 61, 680–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Liu, J.; Li, Y.; Liu, L.; Zhang, X.; Ma, C.Y.; Hua, S.C.; Yang, M.; Yuan, Q. microRNA-1274a, a modulator of sorafenib induced a disintegrin and metalloproteinase 9 (ADAM9) down-regulation in hepatocellular carcinoma. FEBS Lett. 2011, 585, 1828–1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Liu, W.; Ding, R.; Xiong, L.; Dou, R.; Zhang, Y.; Guo, Z. Comprehensive expression profiling and functional network analysis of p53-regulated microRNAs in HepG2 cells treated with doxorubicin. PLoS ONE 2016, 11, e0149227. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhao, R.H.; Tseng, K.F.; Li, K.P.; Lu, Z.G.; Liu, Y.; Han, K.; Gan, Z.H.; Lin, S.C.; Hu, H.Y.; et al. Sirolimus induces apoptosis and reverses multidrug resistance in human osteosarcoma cells in vitro via increasing microRNA-34b expression. Acta Pharmacol. Sin. 2016, 37, 519–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Yan, S.; Zhou, T.; Terada, Y.; Erikson, R.L. The MAP kinase pathway is required for entry into mitosis and cell survival. Oncogene 2004, 23, 763–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borovski, T.; Vellinga, T.T.; Laoukili, J.; Santo, E.E.; Fatrai, S.; van Schelven, S.; Verheem, A.; Marvin, D.L.; Ubink, I.; Borel Rinkes, I.H.M.; et al. Inhibition of RAF1 kinase activity restores apicobasal polarity and impairs tumour growth in human colorectal cancer. Gut 2017, 66, 1106–1115. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.J.; Lai, J.C.; Thike, A.A.; Lim, J.C.; Tan, S.Y.; Koh, V.C.; Lim, T.H.; Bay, B.H.; Tan, M.H.; Tan, P.H. Novel genetic aberrations in breast phyllodes tumours: Comparison between prognostically distinct groups. Breast Cancer Res. Treat. 2014, 145, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.H.; Hang, J.B.; Hu, J.A.; Gao, B.L. RAF1-MEK1-ERK/AKT axis may confer NSCLC cell lines resistance to erlotinib. Int. J. Clin. Exp. Pathol. 2013, 6, 1493–1504. [Google Scholar] [PubMed]
- Wang, F.; Jiang, C.; Sun, Q.; Yan, F.; Wang, L.; Fu, Z.; Liu, T.; Hu, F. miR-195 is a key regulator of Raf1 in thyroid cancer. Onco Targets Ther. 2015, 8, 3021–3028. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, A.; Ho, M. Role of sorafenib in the treatment of advanced hepatocellular carcinoma: An update. Hepatol. Res. 2013, 43, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.X.; Yu, Y.Y.; Zhou, A.Q.; Zhu, J.L.; Luo, L.N.; Chen, W.Q.; Hu, L.; Chen, G.X. Yangzheng Sanjie decoction regulates proliferation and apoptosis of gastric cancer cells by enhancing let-7a expression. World J. Gastroenterol. 2017, 23, 5538–5548. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Gong, W.; Liu, S.; Li, Q.; Guo, M.; Wang, J.; Wang, S.; Chen, N.; Wang, Y.; Liu, Q.; et al. Ibrutinib targets microRNA-21 in multiple myeloma cells by inhibiting NF-κB and STAT3. Tumour Biol. 2018, 40. [Google Scholar] [CrossRef] [PubMed]
- Cutando, A.; Aneiros-Fernández, J.; Aneiros-Cachaza, J.; Arias-Santiago, S. Melatonin and cancer: Current knowledge and its application to oral cavity tumours. J. Oral Pathol. Med. 2011, 40, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.M.; Blask, D.E.; Xiang, S.; Yuan, L.; Mao, L.; Dauchy, R.T.; Dauchy, E.M.; Frasch, T.; Duplesis, T. Melatonin and associated signaling pathways that control normal breast epithelium and breast cancer. J. Mammary Gland Biol. Neoplasia 2011, 16, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Winczyk, K.; Pawlikowski, M.; Lawnicka, H.; Kunert-Radek, J.; Spadoni, G.; Tarzia, G.; Karasek, M. Effects of melatonin and melatonin receptors ligand N-[(4-methoxy-1H-indol-2-yl)methyl]propanamide on murine Colon 38 cancer growth in vitro and in vivo. Neuro Endocrinol. Lett. 2002, 23 (Suppl. 1), 50–54. [Google Scholar] [PubMed]
- Chen, C.Y.; Chen, C.C.; Shieh, T.M.; Hsueh, C.; Wang, S.H.; Leu, Y.L.; Lian, J.H.; Wang, T.H. Corylin suppresses hepatocellular carcinoma progression via the inhibition of epithelial-mesenchymal transition, mediated by long noncoding RNA GAS5. Int. J. Mol. Sci. 2018, 19, 380. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.H.; Yu, C.C.; Lin, Y.S.; Chen, T.C.; Yeh, C.T.; Liang, K.H.; Shieh, T.M.; Chen, C.Y.; Hsueh, C. Long noncoding RNA CPS1-IT1 suppresses the metastasis of hepatocellular carcinoma by regulating HIF-1α activity and inhibiting epithelial-mesenchymal transition. Oncotarget 2016, 7, 43588–43603. [Google Scholar] [CrossRef] [PubMed] [Green Version]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.-H.; Hsueh, C.; Chen, C.-C.; Li, W.-S.; Yeh, C.-T.; Lian, J.-H.; Chang, J.-L.; Chen, C.-Y. Melatonin Inhibits the Progression of Hepatocellular Carcinoma through MicroRNA Let7i-3p Mediated RAF1 Reduction. Int. J. Mol. Sci. 2018, 19, 2687. https://doi.org/10.3390/ijms19092687
Wang T-H, Hsueh C, Chen C-C, Li W-S, Yeh C-T, Lian J-H, Chang J-L, Chen C-Y. Melatonin Inhibits the Progression of Hepatocellular Carcinoma through MicroRNA Let7i-3p Mediated RAF1 Reduction. International Journal of Molecular Sciences. 2018; 19(9):2687. https://doi.org/10.3390/ijms19092687
Chicago/Turabian StyleWang, Tong-Hong, Chuen Hsueh, Chin-Chuan Chen, Wan-Syuan Li, Chau-Ting Yeh, Jang-Hau Lian, Junn-Liang Chang, and Chi-Yuan Chen. 2018. "Melatonin Inhibits the Progression of Hepatocellular Carcinoma through MicroRNA Let7i-3p Mediated RAF1 Reduction" International Journal of Molecular Sciences 19, no. 9: 2687. https://doi.org/10.3390/ijms19092687
APA StyleWang, T. -H., Hsueh, C., Chen, C. -C., Li, W. -S., Yeh, C. -T., Lian, J. -H., Chang, J. -L., & Chen, C. -Y. (2018). Melatonin Inhibits the Progression of Hepatocellular Carcinoma through MicroRNA Let7i-3p Mediated RAF1 Reduction. International Journal of Molecular Sciences, 19(9), 2687. https://doi.org/10.3390/ijms19092687