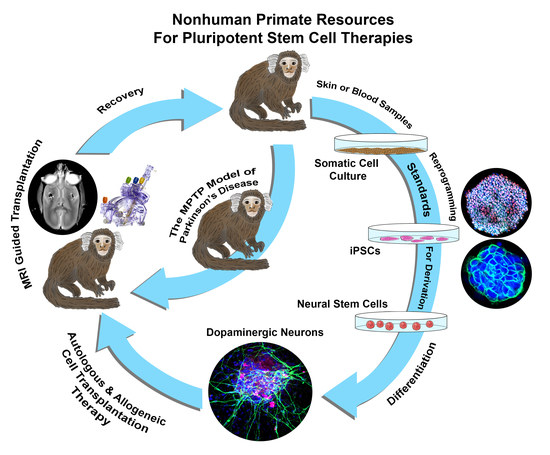
Standards for Deriving Nonhuman Primate-Induced Pluripotent Stem Cells, Neural Stem Cells and Dopaminergic Lineage
Abstract
:1. Introduction
2. Results
2.1. Derivation of Marmoset iPSC Using Human Reprogramming Factors
2.2. Characterization and Banking of the NHP iPSCs
2.3. The Q-PCR Scorecard Standard to Select Pluripotent iPSC Lines
2.4. Derivation of Neural Stem Cells (NSCs) from iPSCs and Their Differentiation into Dopaminergic Lineage
3. Discussions
4. Materials & Methods
4.1. Generation of Marmoset iPS Cells from Skin Biopsy
- The biopsies were transported to our cell culture facility, minced into small pieces (1 mm), and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% of fetal bovine serum (FBS, Hyclone, ThermoFisher Scientific, Waltham, MA, USA) and 1% of penicillin/streptomycin (Gibco, ThermoFisher Scientific, Waltham, MA, USA).
- The fibroblast cultures were usually confluent after 2–3 weeks, passaged once, and tested for the presence of mycoplasma using Hoechst 33258 DNA staining and the mycoplasma PCR detection Kit (Sigma-Aldrich, St. Louis, MO, USA).
- Quality control checks also confirmed freedom from advantageous agents and viability superior to 80%. The selected fibroblasts were expanded for two to five passages to freeze a stock and to transfect with the reprogramming factors.
- Nucleofection of the reprogramming factors: the fibroblasts passage numbers 3 to 6 were trypsinized and five hundred thousand fibroblasts were nucleofected with human episomal plasmids (pCXLE-hOCT3/4-shp53-F, pCXLE-hSK, pCXLE-hUL, and pCXWB-EBNA1; Addgene plasmid #27077, 27078, 27080, and 37624) using the Amaxa Nucleofector device and neonatal human dermal fibroblasts (NHDF) Nucleofector kit Program U-023 (Lonza, Walkersille, MD, USA).
- Nucleofected fibroblasts were transferred onto an irradiated mouse embryonic fibroblasts (MEF) feeder and incubated for three days in DMEM/F12, 20% KnockOut Serum Replacement, 2 mM l-glutamine, MEM Non-Essential Amino Acids Solution, β-Mercaptoethanol (all from Gibco, ThermoFisher Scientific), and 10 ng/mL basic fibroblast growth factor (bFGF, Stemgent, Beltsville, MD, USA). The cultures were maintained for five weeks until the colonies emerged.
- When human embryonic stem cell like colonies appeared, they were isolated, dissociated by pipetting, and transferred into a single well on an irradiated MEF feeder. The H9 hESCs were also cultured on feeder layer as we previously reported [43].
- Validated and authenticated iPSC lines were scaled up to generate 100 vials of 5 to 10 million cell/vial cryopreserved as a seed master cell bank (MCB) for each iPSC line. The iPSCs were cryopreserved in their growth media supplemented with 40% serum and 10% dimethylsulfoxide (DMSO) as cryprotectant.
- One vial of the iPSCs taken from the MCB is used to create a working cell bank.
4.2. Complementary DNA Synthesis, Q-PCR Analysis, and TaqMan Q-PCR Analysis
4.3. Embryoid Body (EB) Formation and Scorecard Analysis
- CJ01 and CJ02 iPSC lines and the H9 hESC line were cultured confluent in six-well plates and the colonies were collected using cell scrapper and cultured in EB media (DMEM/F12, 20% KnockOut Serum Replacement, 1 mM Non-Essential Amino Acids Solution, 55 μM β-Mercaptoethanol).
- The cell suspensions were plated on non-adhesive six-well plates for seven days. Media was changed every two days.
- On the seventh day, the EB’s suspension was collected and RNA was extracted using RNeasy-Plus Mini-kit (QIAGEN).
- Synthesize of the cDNAs was performed using the SuperScript IV First-Strand Synthesis system (Invitrogen).
- The cDNAs were used in the TaqMan hPSC Scorecard Panel (ThermoFisher Scientific) with TaqMan Fast Advanced Master Mix (ThermoFisher Scientific) as per the manufacturer’s protocol.
- Data showing the relative levels of self-renewal genes, mesodermal genes, ectoderm, and endodermal genes are analyzed using the cloud-based software provided with the Scorecard.
- Analyzed data shown a pass or fail results of the pluripotency test, indicating whether the cell line is pluripotent or biased to any of the germ layers.
4.4. Generation of NSCs and Differentiation into Dopaminergic Neurons:
4.5. Immunocytochemistry
4.6. Alkaline Phosphatase (AP) Staining
4.7. Karyotyping
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Mishra, A.; Qiu, Z.; Farnsworth, S.; Tardif, S.D.; Hornsby, P.J. Nonhuman primate induced pluripotent stem cells in regenerative medicine. Stem Cells Int. 2012, 2012, 767195. [Google Scholar] [CrossRef] [PubMed]
- Navara, C.S.; Hornecker, J.; Grow, D.; Chaudhari, S.; Hornsby, P.J.; Ichida, J.K.; Eggan, K.; McCarrey, J.R. Derivation of induced pluripotent stem cells from the baboon: A nonhuman primate model for preclinical testing of stem cell therapies. Cell Reprogram. 2013, 15, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Wunderlich, S.; Kircher, M.; Vieth, B.; Haase, A.; Merkert, S.; Beier, J.; Gohring, G.; Glage, S.; Schambach, A.; Curnow, E.C.; et al. Primate iPS cells as tools for evolutionary analyses. Stem Cell Res. 2014, 12, 622–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimozawa, N.; Ono, R.; Shimada, M.; Shibata, H.; Takahashi, I.; Inada, H.; Takada, T.; Nosaka, T.; Yasutomi, Y. Cynomolgus monkey induced pluripotent stem cells established by using exogenous genes derived from the same monkey species. Differentiation 2013, 85, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Marchetto, M.C.; Narvaiza, I.; Denli, A.M.; Benner, C.; Lazzarini, T.A.; Nathanson, J.L.; Paquola, A.C.; Desai, K.N.; Herai, R.H.; Weitzman, M.D.; et al. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 2013, 503, 525–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hrvoj-Mihic, B.; Marchetto, M.C.; Gage, F.H.; Semendeferi, K.; Muotri, A.R. Novel tools, classic techniques: Evolutionary studies using primate pluripotent stem cells. Biol. Psychiatry 2014, 75, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Wunderlich, S.; Haase, A.; Merkert, S.; Beier, J.; Schwanke, K.; Schambach, A.; Glage, S.; Gohring, G.; Curnow, E.C.; Martin, U. Induction of pluripotent stem cells from a cynomolgus monkey using a polycistronic simian immunodeficiency virus-based vector, differentiation toward functional cardiomyocytes, and generation of stably expressing reporter lines. Cell Reprogram. 2012, 14, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Ben-Nun, I.F.; Montague, S.C.; Houck, M.L.; Tran, H.T.; Garitaonandia, I.; Leonardo, T.R.; Wang, Y.C.; Charter, S.J.; Laurent, L.C.; Ryder, O.A.; et al. Induced pluripotent stem cells from highly endangered species. Nat. Methods 2011, 8, 829–831. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, F.; Yong, J.; Zhang, P.; Hou, P.; Li, H.; Jiang, W.; Cai, J.; Liu, M.; Cui, K.; et al. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell 2008, 3, 587–590. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A.; Kalishman, J.; Golos, T.G.; Durning, M.; Harris, C.P.; Hearn, J.P. Pluripotent cell lines derived from common marmoset (Callithrix jacchus) blastocysts. Biol. Reprod. 1996, 55, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Nii, T.; Marumoto, T.; Kawano, H.; Yamaguchi, S.; Liao, J.; Okada, M.; Sasaki, E.; Miura, Y.; Tani, K. Analysis of essential pathways for self-renewal in common marmoset embryonic stem cells. FEBS Open Bio 2014, 4, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, E.; Hanazawa, K.; Kurita, R.; Akatsuka, A.; Yoshizaki, T.; Ishii, H.; Tanioka, Y.; Ohnishi, Y.; Suemizu, H.; Sugawara, A.; et al. Establishment of novel embryonic stem cell lines derived from the common marmoset (Callithrix jacchus). Stem Cells 2005, 23, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Iwai, H.; Shimada, H.; Nishimura, S.; Kobayashi, Y.; Itakura, G.; Hori, K.; Hikishima, K.; Ebise, H.; Negishi, N.; Shibata, S.; et al. Allogeneic Neural Stem/Progenitor Cells Derived from Embryonic Stem Cells Promote Functional Recovery after Transplantation into Injured Spinal Cord of Nonhuman Primates. Stem Cells Transl. Med. 2015, 4, 708–719. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Matsumura, Y.; Sato, Y.; Okada, A.; Morizane, A.; Okamoto, S.; Hong, H.; Nakagawa, M.; Tanabe, K.; Tezuka, K.; et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 2011, 8, 409–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, F.J.; Goldmann, J.; Loser, P.; Loring, J.F. A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell 2010, 6, 412–414. [Google Scholar] [CrossRef] [PubMed]
- Daley, G.Q.; Lensch, M.W.; Jaenisch, R.; Meissner, A.; Plath, K.; Yamanaka, S. Broader implications of defining standards for the pluripotency of iPSCs. Cell Stem Cell 2009, 4, 200–201. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. Putting stem cells to the test. Nat. Med. 2010, 16, 1354–1357. [Google Scholar] [CrossRef] [PubMed]
- Muller, F.J.; Schuldt, B.M.; Williams, R.; Mason, D.; Altun, G.; Papapetrou, E.P.; Danner, S.; Goldmann, J.E.; Herbst, A.; Schmidt, N.O.; et al. A bioinformatic assay for pluripotency in human cells. Nat. Methods 2011, 8, 315–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bock, C.; Kiskinis, E.; Verstappen, G.; Gu, H.; Boulting, G.; Smith, Z.D.; Ziller, M.; Croft, G.F.; Amoroso, M.W.; Oakley, D.H.; et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 2011, 144, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Tsankov, A.M.; Akopian, V.; Pop, R.; Chetty, S.; Gifford, C.A.; Daheron, L.; Tsankova, N.M.; Meissner, A. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat. Biotechnol. 2015, 33, 1182–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daadi, M.M.; Weiss, S. Generation of tyrosine hydroxylase-producing neurons from precursors of the embryonic and adult forebrain. J. Neurosci. 1999, 19, 4484–4497. [Google Scholar] [CrossRef] [PubMed]
- Stroke Therapy Academic Industry Roundtable (STAIR). Recommendations for standards regarding preclinical neuroprotective and restorative drug development. Stroke 1999, 30, 2752–2758. [Google Scholar] [CrossRef]
- Tesar, P.J.; Chenoweth, J.G.; Brook, F.A.; Davies, T.J.; Evans, E.P.; Mack, D.L.; Gardner, R.L.; McKay, R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007, 448, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Izpisua Belmonte, J.C. Dynamic Pluripotent Stem Cell States and Their Applications. Cell Stem Cell 2015, 17, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, B.; Xu, J.; Wang, J.; Wu, J.; Shi, C.; Xu, Y.; Dong, J.; Wang, C.; Lai, W.; et al. Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell 2017, 169, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Vermilyea, S.C.; Guthrie, S.; Meyer, M.; Smuga-Otto, K.; Braun, K.; Howden, S.; Thomson, J.A.; Zhang, S.C.; Emborg, M.E.; Golos, T.G. Induced Pluripotent Stem Cell-Derived Dopaminergic Neurons from Adult Common Marmoset Fibroblasts. Stem Cells Dev. 2017, 26, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Torrez, L.B.; Perez, Y.; Yang, J.; Zur Nieden, N.I.; Klassen, H.; Liew, C.G. Derivation of neural progenitors and retinal pigment epithelium from common marmoset and human pluripotent stem cells. Stem Cells Int. 2012, 2012, 417865–417874. [Google Scholar] [CrossRef] [PubMed]
- Shimada, H.; Okada, Y.; Ibata, K.; Ebise, H.; Ota, S.; Tomioka, I.; Nomura, T.; Maeda, T.; Kohda, K.; Yuzaki, M.; et al. Efficient derivation of multipotent neural stem/progenitor cells from non-human primate embryonic stem cells. PLoS ONE 2012, 7, e49469. [Google Scholar] [CrossRef] [PubMed]
- Farnsworth, S.L.; Qiu, Z.; Mishra, A.; Hornsby, P.J. Directed neural differentiation of induced pluripotent stem cells from non-human primates. Exp. Biol. Med. 2013, 238, 276–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomioka, I.; Maeda, T.; Shimada, H.; Kawai, K.; Okada, Y.; Igarashi, H.; Oiwa, R.; Iwasaki, T.; Aoki, M.; Kimura, T.; et al. Generating induced pluripotent stem cells from common marmoset (Callithrix jacchus) fetal liver cells using defined factors, including Lin28. Genes Cells 2010, 15, 959–969. [Google Scholar] [CrossRef] [PubMed]
- Debowski, K.; Warthemann, R.; Lentes, J.; Salinas-Riester, G.; Dressel, R.; Langenstroth, D.; Gromoll, J.; Sasaki, E.; Behr, R. Non-viral generation of marmoset monkey iPS cells by a six-factor-in-one-vector approach. PLoS ONE 2015, 10, e0118424. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, A.; Hemmer, K.; Bernemann, I.; Gohring, G.; Pogozhykh, O.; Figueiredo, C.; Glage, S.; Schambach, A.; Schwamborn, J.C.; Blasczyk, R.; et al. Induced pluripotent stem cells generated from adult bone marrow-derived cells of the nonhuman primate (Callithrix jacchus) using a novel quad-cistronic and excisable lentiviral vector. Cell Reprogram. 2012, 14, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Kohda, K.; Ibata, K.; Kohyama, J.; Akamatsu, W.; Yuzaki, M.; Okano, H.J.; Sasaki, E.; Okano, H. Reprogramming non-human primate somatic cells into functional neuronal cells by defined factors. Mol. Brain 2014, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Mishra, A.; Li, M.; Farnsworth, S.L.; Guerra, B.; Lanford, R.E.; Hornsby, P.J. Marmoset induced pluripotent stem cells: Robust neural differentiation following pretreatment with dimethyl sulfoxide. Stem Cell Res. 2015, 15, 141–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reubinoff, B.E.; Itsykson, P.; Turetsky, T.; Pera, M.F.; Reinhartz, E.; Itzik, A.; Ben-Hur, T. Neural progenitors from human embryonic stem cells. Nat. Biotechnol. 2001, 19, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.C.; Wernig, M.; Duncan, I.D.; Brustle, O.; Thomson, J.A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001, 19, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Bain, G.; Kitchens, D.; Yao, M.; Huettner, J.E.; Gottlieb, D.I. Embryonic stem cells express neuronal properties in vitro. Dev. Biol. 1995, 168, 342–357. [Google Scholar] [CrossRef] [PubMed]
- Okabe, S.; Forsberg-Nilsson, K.; Spiro, A.C.; Segal, M.; McKay, R.D. Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech. Dev. 1996, 59, 89–102. [Google Scholar] [CrossRef]
- Ying, Q.L.; Stavridis, M.; Griffiths, D.; Li, M.; Smith, A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003, 21, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Daadi, M.M.; Maag, A.L.; Steinberg, G.K. Adherent self-renewable human embryonic stem cell-derived neural stem cell line: Functional engraftment in experimental stroke model. PLoS ONE 2008, 3, e1644. [Google Scholar] [CrossRef] [PubMed]
- Daadi, M.M.; Grueter, B.A.; Malenka, R.C.; Redmond, D.E., Jr.; Steinberg, G.K. Dopaminergic neurons from midbrain-specified human embryonic stem cell-derived neural stem cells engrafted in a monkey model of Parkinson’s disease. PLoS ONE 2012, 7, e41120. [Google Scholar] [CrossRef] [PubMed]
- Kriks, S.; Shim, J.W.; Piao, J.; Ganat, Y.M.; Wakeman, D.R.; Xie, Z.; Carrillo-Reid, L.; Auyeung, G.; Antonacci, C.; Buch, A.; et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 2011, 480, 547–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, G.; Hong, H.; Torres, A.; Malloy, K.E.; Choudhury, G.R.; Kim, J.; Daadi, M.M. Standards for Deriving Nonhuman Primate-Induced Pluripotent Stem Cells, Neural Stem Cells and Dopaminergic Lineage. Int. J. Mol. Sci. 2018, 19, 2788. https://doi.org/10.3390/ijms19092788
Yang G, Hong H, Torres A, Malloy KE, Choudhury GR, Kim J, Daadi MM. Standards for Deriving Nonhuman Primate-Induced Pluripotent Stem Cells, Neural Stem Cells and Dopaminergic Lineage. International Journal of Molecular Sciences. 2018; 19(9):2788. https://doi.org/10.3390/ijms19092788
Chicago/Turabian StyleYang, Guang, Hyenjong Hong, April Torres, Kristen E. Malloy, Gourav R. Choudhury, Jeffrey Kim, and Marcel M. Daadi. 2018. "Standards for Deriving Nonhuman Primate-Induced Pluripotent Stem Cells, Neural Stem Cells and Dopaminergic Lineage" International Journal of Molecular Sciences 19, no. 9: 2788. https://doi.org/10.3390/ijms19092788
APA StyleYang, G., Hong, H., Torres, A., Malloy, K. E., Choudhury, G. R., Kim, J., & Daadi, M. M. (2018). Standards for Deriving Nonhuman Primate-Induced Pluripotent Stem Cells, Neural Stem Cells and Dopaminergic Lineage. International Journal of Molecular Sciences, 19(9), 2788. https://doi.org/10.3390/ijms19092788