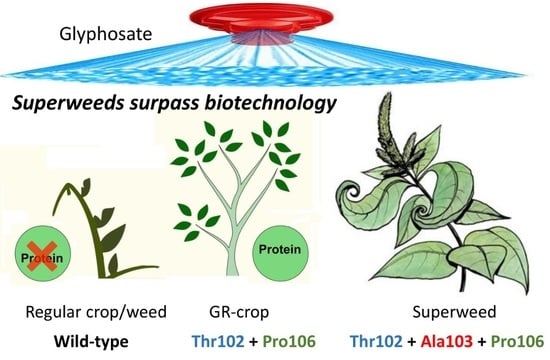
The Triple Amino Acid Substitution TAP-IVS in the EPSPS Gene Confers High Glyphosate Resistance to the Superweed Amaranthus hybridus
Abstract
:1. Introduction
2. Results
2.1. Shikimic Acid Accumulation and Genotyping
2.2. Dose–response Assays
2.3. 14C-Glyphosate Absorption and Translocation
2.4. Glyphosate Metabolism
2.5. EPSPS Enzyme Activity Assays
2.6. EPSPS Copy Number and Gene Expression
2.7. Mutations in the EPSPS Coding Sequence
2.8. Structural Modeling
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Fast Screening of Resistance via Shikimic Acid Accumulation
4.3. Species Identification
4.4. Dose–response Assays
4.5. 14C-Glyphosate Absorption and Translocation
4.6. 14C-Glyphosate Visualization
4.7. Glyphosate Metabolism
4.8. EPSPS Enzyme Activity Assays
4.9. EPSPS Copy Number and Gene Expression
4.10. Mutations in the EPSPS Coding Sequence
4.11. Structural Modeling
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Domínguez-Mendez, R.; Alcántara-de la Cruz, R.; Rojano-Delgado, A.M.; Silveira, H.M.; Portugal, J.; Cruz-Hipolito, H.E.; De Prado, R. Stacked traits conferring multiple resistance to imazamox and glufosinate in soft wheat. Pest Manag. Sci. 1979, 75, 648–657. [Google Scholar]
- Green, J.M. The rise and future of glyphosate and glyphosate-resistant crops. Pest Manag. Sci. 2018, 74, 1035–1039. [Google Scholar] [CrossRef]
- Silveira, H.M.; Langaro, A.C.; Alcántara-de la Cruz, R.A.; Sediyama, T.; Silva, A.A. Glyphosate efficacy on sourgrass biotypes with suspected resistance collected in GR-crop fields. Acta Sci. Agron. 2018, 40, 35120. [Google Scholar] [CrossRef] [Green Version]
- Bonny, S. Genetically modified herbicide-tolerant crops, weeds, and herbicides: Overview and impact. Environ. Manage. 2016, 57, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Bain, C.; Selfa, T.; Dandachi, T.; Velardi, S. ‘Superweeds’ or ‘survivors’? Framing the problem of glyphosate resistant weeds and genetically engineered crops. J. Rural Stud. 2017, 51, 211–221. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Devos, Y.; Beckie, H.J.; Owen, M.D.K.; Tillie, P.; Messéan, A.; Kudsk, P. Integrated weed management systems with herbicide-tolerant crops in the European Union: Lessons learnt from home and abroad. Crit. Rev. Biotechnol. 2017, 37, 459–475. [Google Scholar] [CrossRef]
- Heap, I.; Duke, S.O. Overview of glyphosate-resistant weeds worldwide. Pest Manag. Sci. 2018, 74, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Sammons, R.D.; Gaines, T.A. Glyphosate resistance: State of knowledge. Pest Manag. Sci. 2014, 70, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Duke, S.O. The history and current status of glyphosate. Pest Manag. Sci. 2018, 74, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Gaines, T.A.; Zhang, W.; Wang, D.; Bukun, B.; Chisholm, S.T.; Shaner, D.L.; Nissen, S.J.; Patzoldt, W.L.; Tranel, P.J.; Culpepper, A.S.; et al. Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Natl. Acad. Sci. USA 2010, 107, 1029–1034. [Google Scholar] [CrossRef]
- Shaner, D.L.; Nadler-Hassar, T.; Henry, W.B.; Koger, C.H. A rapid in vivo shikimate accumulation assay with excised leaf discs. Weed Sci. 2005, 53, 769–774. [Google Scholar] [CrossRef]
- Duhoux, A.; Carrère, S.; Duhoux, A.; Délye, C. Transcriptional markers enable identification of rye-grass (Lolium sp.) plants with non-target-site-based resistance to herbicides inhibiting acetolactate-synthase. Plant Sci. 2017, 257, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Alcántara-de la Cruz, R.; Fernández-Moreno, P.T.; Ozuna, C.V.; Rojano-Delgado, A.M.; Cruz-Hipolito, H.E.; Domínguez-Valenzuela, J.A.; Barro, F.; De Prado, R. Target and non-target site mechanisms developed by glyphosate-resistant Hairy beggarticks (Bidens pilosa L.) populations from Mexico. Front. Plant Sci. 2016, 7, 1492. [Google Scholar]
- Ge, X.; D’Avignon, D.A.; Ackerman, J.J.; Sammons, R.D. Rapid vacuolar sequestration: The horseweed glyphosate resistance mechanism. Pest Manag. Sci. 2010, 66, 345–348. [Google Scholar] [CrossRef]
- Ghanizadeh, H.; Harrington, K.C. Non-target site mechanisms of resistance to herbicides. Crit. Rev. Plant. Sci. 2017, 36, 24–34. [Google Scholar] [CrossRef]
- Amaro-Blanco, I.; Fernández-Moreno, P.T.; Osuna-Ruiz, M.D.; Bastida, F.; De Prado, R. Mechanisms of glyphosate resistance and response to alternative herbicide-based management in populations of the three Conyza species introduced in southern Spain. Pest Manag. Sci. 2018, 74, 1925–1937. [Google Scholar] [CrossRef]
- Gaines, T.A.; Patterson, E.L.; Neve, P. Molecular mechanisms of adaptive evolution revealed by global selection for glyphosate resistance. New Phytol. 2019. (In Press) [Google Scholar] [CrossRef]
- Vitta, J.I.; Faccini, D.E.; Nisensohn, L.A. Control of Amaranthus quitensis in soybean crops in Argentina: An alternative to reduce herbicide use. Crop Prot. 2000, 19, 511–513. [Google Scholar] [CrossRef]
- Faccini, D.; Vitta, J.I. Germination characteristics of Amaranthus quitensis as affected by seed production date and duration of burial. Weed Res. 2005, 45, 371–378. [Google Scholar] [CrossRef]
- Dellaferrera, I.; Cortés, E.; Panigo, E.; De Prado, R.; Chritoffoleti, P.; Perreta, M. First report of Amaranthus hybridus with multiple resistance to 2,4-D, dicamba, and glyphosate. Agronomy 2018, 8, 140. [Google Scholar] [CrossRef]
- Varanasi, V.K.; Godar, A.S.; Currie, R.S.; Dille, A.J.; Thompson, C.R.; Stahlman, P.W.; Jugulam, M. Field-evolved resistance to four modes of action of herbicides in a single kochia (Kochia scoparia L. Schrad.) population. Pest Manag. Sci. 2015, 71, 1207–1212. [Google Scholar] [CrossRef]
- Shergill, L.S.; Bish, M.D.; Jugulam, M.; Bradley, K.W. Molecular and physiological characterization of six-way resistance in an Amaranthus tuberculatus var. rudis biotype from Missouri. Pest Manag. Sci. 2018, 74, 2688–2698. [Google Scholar] [CrossRef]
- Tahmasebi, B.K.; Alcántara-de la Cruz, R.; Alcántara, E.; Torra, J.; Domínguez-Valenzuela, J.A.; Cruz-Hipólito, H.E.; Rojano-Delgado, A.M.; De Prado, R. Multiple resistance evolution in bipyridylium-resistant Epilobium ciliatum after recurrent selection. Front. Plant Sci. 2018, 9, 695. [Google Scholar] [CrossRef]
- Nandula, V.K.; Wright, A.A.; Bond, J.A.; Ray, J.D.; Eubank, T.W.; Molin, W.T. EPSPS amplification in glyphosate-resistant spiny amaranth (Amaranthus spinosus): A case of gene transfer via interspecific hybridization from glyphosate-resistant Palmer amaranth (Amaranthus palmeri). Pest Manag. Sci. 2014, 70, 1902–1909. [Google Scholar] [CrossRef]
- Lorentz, L.; Gaines, T.A.; Nissen, S.J.; Westra, P.; Strek, H.J.; Dehne, H.W.; Ruiz-Santaella, J.P.; Beffa, R. Characterization of glyphosate resistance in Amaranthus tuberculatus populations. J. Agric. Food Chem. 2014, 62, 8134–8142. [Google Scholar] [CrossRef]
- Garcia-del Rosal, M.J.; Bracamonte, E.; Fernandez-Moreno, P.T.; Alcantara-de la Cruz, R.; De Prado, R. A novel amino acid substitution Ala-103-Val in EPSPS has been found in glyphosate resistant A. hybridus. In Proceedings of the 58th WSSA Annual Meeting, Arlintong, VA, USA, 29 January–1 February 2018. [Google Scholar]
- Wright, A.A.; Molin, W.T.; Nandula, V.K. Distinguishing between weedy Amaranthus species based on intron 1 sequences from the 5-enolpyruvylshikimate-3-phosphate synthase gene. Pest Manag. Sci. 2016, 72, 2347–2354. [Google Scholar] [CrossRef]
- Palma-Bautista, C.; Torra, J.; Garcia, M.J. Reduced absorption and impaired translocation endows glyphosate resistance in Amaranthus palmeri harvested in glyphosate-resistant soybean from Argentina. J. Agric. Food Chem. 2019, 67, 1052–1060. [Google Scholar] [CrossRef]
- Perotti, V.E.; Larran, A.S.; Palmeari, V.E.; Martinatto, A.K.; Alvarez, C.E.; Tuesca, D.; Permingeat, H.R. A novel triple amino acid substitution in the EPSPS found in a high-level glyphosate resistant Amaranthus hybridus population from Argentina. Pest Manag. Sci. 2019, 75, 242–1251. [Google Scholar] [CrossRef]
- González-Torralva, F.; Cruz-Hipolito, H.; Bastida, F.; Mülleder, N.; Smeda, R.J.; De Prado, R. Differential susceptibility to glyphosate among the Conyza weed species in Spain. J. Agric. Food Chem. 2010, 58, 4361–4366. [Google Scholar] [CrossRef]
- Vila-Aiub, M.M.; Balbil, M.C.; Distéfano, A.J. Glyphosate resistance in perennial Sorghum halepense (Johnsongrass), endowed by reduced glyphosate translocation and leaf uptake. Pest Manag. Sci. 2012, 68, 430–436. [Google Scholar] [CrossRef]
- Binimelis, R.; Pengue, W.; Monterroso, I. “Transgenic treadmill”: Responses to the emergence and spread of glyphosate-resistant johnsongrass in Argentina. Geoforum 2009, 40, 623–633. [Google Scholar] [CrossRef]
- Preston, C.; Wakelin, A.M. Resistance to glyphosate from altered herbicide translocation patterns. Pest Manag. Sci. 2008, 64, 372–376. [Google Scholar] [CrossRef]
- Ge, X.; D’Avignon, D.A.; Ackerman, J.J.; Collavo, A.; Sattin, M.; Ostrander, E.L.; Hall, E.L.; Sammons, R.D.; Preston, C. Vacuolar glyphosate-sequestration correlates with glyphosate resistance in Ryegrass (Lolium spp.) from Australia, South America, and Europe: A 31P NMR investigation. J. Agric. Food Chem. 2012, 60, 1243–1250. [Google Scholar] [CrossRef]
- Ge, X.; D’Avignon, D.A.; Ackerman, J.J.; Sammons, R.D. In vivo 31P-nuclear magnetic resonance studies of glyphosate uptake, vacuolar sequestration, and tonoplast pump activity in glyphosate-resistant horseweed. Plant Physiol. 2014, 166, 1255–1268. [Google Scholar] [CrossRef]
- Yu, Q.; Jalaludin, A.; Han, H.; Chen, M.; Sammons, R.D.; Powles, S.B. Evolution of a double amino acid substitution in the 5-enolpyruvylshikimate-3-phosphate synthase in Eleusine indica conferring high-level glyphosate resistance. Plant Physiol. 2015, 167, 1440–1447. [Google Scholar] [CrossRef]
- Nandula, V.K.; Ray, J.D.; Ribeiro, D.N.; Pan, Z.; Reddy, K.N. Glyphosate resistance in tall waterhemp (Amaranthus tuberculatus) from Mississippi is due to both altered target-site and nontarget-site mechanisms. Weed Sci. 2013, 61, 374–383. [Google Scholar] [CrossRef]
- Riggins, C.W.; Peng, Y.H.; Stewart, C.N.; Tranel, P.J. Characterization of de novo transcriptome for waterhemp (Amaranthus tuberculatus) using GS-FLX 454 pyrosequencing and its application for studies of herbicide target-site genes. Pest Manag. Sci. 2010, 66, 1042–1052. [Google Scholar] [CrossRef]
- Bracamonte, E.; Silveira, H.M.; Alcántara-de la Cruz, R.; Domínguez-Valenzuela, J.A.; Cruz-Hipólito, H.E.; De Prado, R. From tolerance to resistance: Mechanisms governing the differential response to glyphosate in Chloris barbata. Pest Manag. Sci. 2018, 74, 1118–1124. [Google Scholar] [CrossRef]
- Healy-Fried, M.L.; Funke, T.; Priestman, M.A.; Han, H.; Schönbrunn, E. Structural basis of glyphosate tolerance resulting from mutations of Pro101 in Escherichia coli 5-enolpyruvylshikimate-3-phosphate synthase. J. Biol. Chem. 2007, 282, 32949–32955. [Google Scholar] [CrossRef]
- Funke, T.; Yang, Y.; Han, H.; Healy-Fried, M.; Olesen, S.; Becker, A.; Schönbrunn, E. Structural basis of glyphosate resistance resulting from the double mutation Thr 97 → Ile and Pro 101 → Ser in 5-enolpyruvylshikimate-3-phosphate synthase from Escherichia coli. J. Biol. Chem. 2009, 284, 9854–9860. [Google Scholar] [CrossRef]
- Eschenburg, S.; Healy, M.; Priestman, M.; Lushington, G.; Schönbrunn, E. How the mutation glycine96 to alanine confers glyphosate insensitivity to 5-enolpyruvyl shikimate-3-phosphate synthase from Escherichia coli. Planta 2002, 216, 129–135. [Google Scholar] [CrossRef]
- Li, J.; Peng, Q.; Han, H.; Nyporko, A.; Kulynych, T.; Yu, Q.; Powles, S. Glyphosate resistance in Tridax procumbens via a novel EPSPS Thr-102-Ser substitution. J. Agric. Food Chem. 2018, 66, 7880–7888. [Google Scholar] [CrossRef]
- Chen, J.; Huang, H.; Zhang, C.; Wei, S.; Huang, Z.; Chen, J.; Wang, X. Mutations and amplification of EPSPS gene confer resistance to glyphosate in goosegrass (Eleusine indica). Planta 2015, 242, 859–868. [Google Scholar] [CrossRef]
- Betts, M.J.; Russell, R.B. Amino acid properties and consequences of substitutions. In Bioinformatics for Geneticists, 1st ed.; Barnes, M.R., Gray, I.C., Eds.; John Wiley & Sons: Chichester, UK, 2003; pp. 289–316. [Google Scholar]
- Rojano-Delgado, A.M.; Ruiz-Jiménez, J.; Castro, M.D.L.; De Prado, R. Determination of glyphosate and its metabolites in plant material by reversed-polarity CE with indirect absorptiometric detection. Electrophoresis 2010, 31, 1423–1430. [Google Scholar] [CrossRef]
- Dayan, F.E.; Owens, D.K.; Corniani, N.; Silva, F.M.L.; Watson, S.B.; Howell, J.´L.; Shaner, D.L. Biochemical markers and enzyme assays for herbicide mode of action and resistance studies. Weed Sci. 2015, 63, 23–63. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar] [CrossRef]
- Venselaar, H.; te Beek, T.A.; Kuipers, R.K.; Hekkelman, M.L.; Vriend, G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinform. 2010, 11, 548. [Google Scholar] [CrossRef]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-response analysis using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef]







| Population | c | d | b | Concentration | RI | p-value |
|---|---|---|---|---|---|---|
| LD50 (g ae ha−1) † | ||||||
| GRH | - | 98.9 | 0.21 | 3503.4 ± 34.7 | 100.6 | < 0.0001 |
| GSH | - | 100.1 | 0.14 | 34.8 ± 2.8 | -- | < 0.0001 |
| GR50 (g ae ha−1) † | ||||||
| GRH | - | 98.9 | 11.37 | 1395.2 ± 164.5 | 83.9 | < 0.0001 |
| GSH | - | 100.0 | 0.26 | 16.6 ± 1.7 | -- | < 0.0001 |
| I50 (µM) ‡ | ||||||
| GRH | 0.10 | 101.7 | 0.52 | 52.8 ± 3.4 | 100.9 | < 0.0001 |
| GSH | 0.84 | 100.9 | 0.63 | 0.52 ± 0.07 | -- | < 0.0001 |
| Population | HAT | Aerial Part | Roots | ||
|---|---|---|---|---|---|
| Glyphosate | Metabolites | Glyphosate | Metabolites | ||
| GRH | 48 | 436.9 ± 2.5 | ND | 47.9 ± 4.6 | ND |
| 96 | 494.3 ± 3.7 | ND | 114.3 ± 5.0 | ND | |
| GSH | 48 | 522.9 ± 8.4 | ND | 83.5 ± 7.0 | ND |
| 96 | 605.0 ± 12.5 | ND | 237.5 ± 7.2 | ND | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García, M.J.; Palma-Bautista, C.; Rojano-Delgado, A.M.; Bracamonte, E.; Portugal, J.; Alcántara-de la Cruz, R.; De Prado, R. The Triple Amino Acid Substitution TAP-IVS in the EPSPS Gene Confers High Glyphosate Resistance to the Superweed Amaranthus hybridus. Int. J. Mol. Sci. 2019, 20, 2396. https://doi.org/10.3390/ijms20102396
García MJ, Palma-Bautista C, Rojano-Delgado AM, Bracamonte E, Portugal J, Alcántara-de la Cruz R, De Prado R. The Triple Amino Acid Substitution TAP-IVS in the EPSPS Gene Confers High Glyphosate Resistance to the Superweed Amaranthus hybridus. International Journal of Molecular Sciences. 2019; 20(10):2396. https://doi.org/10.3390/ijms20102396
Chicago/Turabian StyleGarcía, Maria J., Candelario Palma-Bautista, Antonia M. Rojano-Delgado, Enzo Bracamonte, João Portugal, Ricardo Alcántara-de la Cruz, and Rafael De Prado. 2019. "The Triple Amino Acid Substitution TAP-IVS in the EPSPS Gene Confers High Glyphosate Resistance to the Superweed Amaranthus hybridus" International Journal of Molecular Sciences 20, no. 10: 2396. https://doi.org/10.3390/ijms20102396
APA StyleGarcía, M. J., Palma-Bautista, C., Rojano-Delgado, A. M., Bracamonte, E., Portugal, J., Alcántara-de la Cruz, R., & De Prado, R. (2019). The Triple Amino Acid Substitution TAP-IVS in the EPSPS Gene Confers High Glyphosate Resistance to the Superweed Amaranthus hybridus. International Journal of Molecular Sciences, 20(10), 2396. https://doi.org/10.3390/ijms20102396