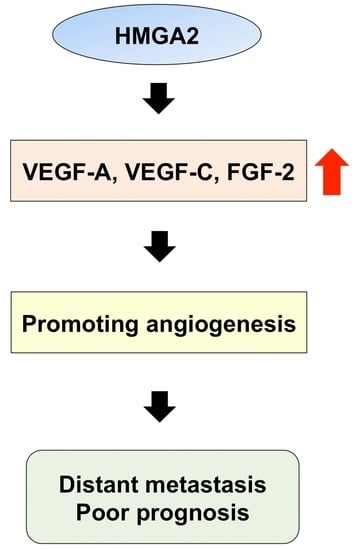
HMGA2 Contributes to Distant Metastasis and Poor Prognosis by Promoting Angiogenesis in Oral Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Results
2.1. Clinical Significance of HMGA2 Expression in OSCC Tissues
2.2. Relationships between the HMGA2 and Survival Time
2.3. Role of HMGA2 in the Transcriptional Control of EMT Markers
2.4. Involvement of HMGA2 in the EMT Phenotype of OSCC Cells
2.5. Relationship between HMGA2 and Angiogenesis-Associated Genes in OSCC Cells
2.6. Expression of HMGA2 and Angiogenesis-Associated Genes in Primary OSCC and Metastatic Tissue
3. Discussion
4. Materials and Methods
4.1. Patients and Tissue Specimens
4.2. Immunohistochemical Staining and Evaluation
4.3. Assessment of Immunohistochemical Staining
4.4. Cell Lines
4.5. Transfection with Small Interfering RNA and Reagent
4.6. Cell Proliferation Analysis
4.7. Scratch Wound Healing Assay
4.8. Matrigel Cell Invasion Assay
4.9. RNA Isolation, Reverse Transcription, and Quantitative PCR
| HMGA2 | forward: 5′-ACCCAGGGGAAGACCCAAA-3′ reverse: 5′-CCTCTTGGCCGTTTTTCT-3′ |
| E-cadherin | forward: 5′-ATTTTTCCCTCGACACCCGAT-3′ reverse: 5′-TCCCAGGCGTAGACCAAGA-3′ |
| Vimentin | forward: 5′-AGTCCACTGAGTACCGGAGAC-3′ reverse: 5′-CATTTCACGCATCTGGCGTTC-3′ |
| Slug | forward: 5′-AAGCATTTCAACGCCTCCAAA-3′ reverse: 5′-GGATCTCTGGTTGTGGTATGACA-3′ |
| FGF-2 | forward: 5′-CACCTATAATTGGTCAAAGTGG-3′ reverse: 5′-CAGAAATTCAGTAGATGTTTCCC-3′ |
| VEGF-A | forward: 5′-CCTCCGAAACCATGAACTTT-3′ reverse: 5′-CCACTTCGTGATGATTCTGC-3′ |
| VEGF-C | forward: 5′-GCCCCAAACCAGTAACAATC-3′ reverse: 5′-GCTGGCAGGGAACGTCTAAT-3′ |
| GAPDH | forward: 5′-CTGGGCTACACTGAGCACC-3′ reverse: 5′-AAGTGGTCGTTGAGGGCAATG-3′ |
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| EMT | Epithelial–mesenchymal transition |
| HMGA2 | High-mobility group A protein 2 |
| OSCC | Oral squamous cell carcinoma |
| qRT-PCR | quantitative RT-PCR |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA-Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Kong, W.; Peng, Y.; Miao, Q.; Mackillop, W.J. Temporal trends in the incidence and survival of cancers of the upper aerodigestive tract in Ontario and the United States. Int. J. Cancer 2009, 125, 2159–2165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasegawa, T.; Tanakura, M.; Takeda, D.; Sakakibara, A.; Akashi, M.; Minamikawa, T.; Komori, T. Risk factors associated with distant metastasis in patients with oral squamous cell carcinoma. Otolaryngol. Head Neck Surg. 2015, 152, 1053–1060. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A. Epithelial plasticity: A common theme in embryonic and cancer cells. Science 2013, 342, 1234850. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Mandal, M.; Myers, J.N.; Lippman, S.M.; Johnson, F.M.; Williams, M.D.; Rayala, S.; Ohshiro, K.; Rosenthal, D.I.; Weber, R.S.; Gallick, G.E.; et al. Epithelial to mesenchymal transition in head and neck squamous carcinoma: Association of Src activation with E-cadherin down-regulation, vimentin expression, and aggressive tumor features. Cancer 2008, 112, 2088–2100. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Olmeda, D.; Cano, A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 2007, 7, 415–428. [Google Scholar] [CrossRef]
- Katsuno, Y.; Lamouille, S.; Derynck, R. TGF-beta signaling and epithelial-mesenchymal transition in cancer progression. Curr. Opin. Oncol. 2013, 25, 76–84. [Google Scholar] [CrossRef]
- Rhim, A.D.; Mirek, E.T.; Aiello, N.M.; Maitra, A.; Bailey, J.M.; McAllister, F.; Reichert, M.; Beatty, G.L.; Rustgi, A.K.; Vonderheide, R.H.; et al. EMT and dissemination precede pancreatic tumor formation. Cell 2012, 148, 349–361. [Google Scholar] [CrossRef] [Green Version]
- Waerner, T.; Alacakaptan, M.; Tamir, I.; Oberauer, R.; Gal, A.; Brabletz, T.; Schreiber, M.; Jechlinger, M.; Beug, H. ILEI: A cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell 2006, 10, 227–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.F.; Yang, X.D.; Lu, M.X.; Sun, G.W.; Wang, Y.X.; Zhang, Y.K.; Pu, Y.M.; Tang, E.Y. Prognostic significance of VEGF immunohistochemical expression in oral cancer: A meta-analysis of the literature. Tumour Biol. 2013, 34, 3165–3171. [Google Scholar] [CrossRef] [PubMed]
- Yadav, L.; Puri, N.; Rastogi, V.; Satpute, P.; Sharma, V. Tumour angiogenesis and angiogenic inhibitors: A review. J. Clin. Diagn. Res. 2015, 9, XE01–XE05. [Google Scholar] [CrossRef] [PubMed]
- Carla, C.; Daris, F.; Cecilia, B.; Francesca, B.; Francesca, C.; Paolo, F. Angiogenesis in head and neck cancer: A review of the literature. J. Oncol. 2012, 2012, 358472. [Google Scholar] [CrossRef]
- Hammond, S.M.; Sharpless, N.E. HMGA2, microRNAs, and stem cell aging. Cell 2008, 135, 1013–1016. [Google Scholar] [CrossRef] [PubMed]
- Fusco, A.; Fedele, M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer 2007, 7, 899–910. [Google Scholar] [CrossRef]
- Mahajan, A.; Liu, Z.; Gellert, L.; Zou, X.; Yang, G.; Lee, P.; Yang, X.; Wei, J.J. HMGA2: A biomarker significantly overexpressed in high-grade ovarian serous carcinoma. Mod. Pathol. 2010, 23, 673–681. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Lv, G.D.; Qin, X.; Gen, Y.H.; Zheng, S.T.; Liu, T.; Lu, X.M. Role of microRNA let-7 and effect to HMGA2 in esophageal squamous cell carcinoma. Mol. Biol. Rep. 2012, 39, 1239–1246. [Google Scholar] [CrossRef]
- Meyer, B.; Loeschke, S.; Schultze, A.; Weigel, T.; Sandkamp, M.; Goldmann, T.; Vollmer, E.; Bullerdiek, J. HMGA2 overexpression in non-small cell lung cancer. Mol. Carcinog. 2007, 46, 503–511. [Google Scholar] [CrossRef]
- Pallante, P.; Sepe, R.; Puca, F.; Fusco, A. High mobility group a proteins as tumor markers. Front. Med. 2015, 2, 15. [Google Scholar] [CrossRef]
- Thuault, S.; Valcourt, U.; Petersen, M.; Manfioletti, G.; Heldin, C.H.; Moustakas, A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J. Cell Biol. 2006, 174, 175–183. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Z.; Xu, C.; Zhou, Z.; Zhu, Z.; You, T. HMGA2 induces transcription factor Slug expression to promote epithelial-to-mesenchymal transition and contributes to colon cancer progression. Cancer Lett. 2014, 355, 130–140. [Google Scholar] [CrossRef]
- Watanabe, S.; Ueda, Y.; Akaboshi, S.; Hino, Y.; Sekita, Y.; Nakao, M. HMGA2 maintains oncogenic RAS-induced epithelial-mesenchymal transition in human pancreatic cancer cells. Am. J. Pathol. 2009, 174, 854–868. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Frost, R.J.A.; Anderson, C.; Zhao, F.; Ma, J.; Yu, B.; Wang, S. Let-7 contributes to diabetic retinopathy but represses pathological ocular angiogenesis. Mol. Cell. Biol. 2017. [Epub ahead of print]. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Deng, S.; Ma, Q.; Zhang, T.; Jia, C.; Zhuo, D.; Yang, F.; Wei, J.; Wang, L.; Dykxhoorn, D.M.; et al. MicroRN-10A* and MicroRNA-21 modulate endothelial progenitor cell senescense via suppressing high-mobility group A2. Circ. Res. 2013, 112, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, J.; Mitoro, A.; Kawashiri, S.; Chada, K.K.; Imai, K. Expression of mesenchyme-specific gene HMGA2 in squamous cell carcinomas of the oral cavity. Cancer Res. 2004, 64, 2024–2029. [Google Scholar] [CrossRef]
- Zhao, X.P.; Zhang, H.; Jiao, J.Y.; Tang, D.X.; Wu, Y.L.; Pan, C.B. Overexpression of HMGA2 promotes tongue cancer metastasis through EMT pathway. J. Transl. Med. 2016, 14, 26. [Google Scholar] [CrossRef]
- Loeschke, S.; Ohlmann, A.K.; Brasen, J.H.; Holst, R.; Warnke, P.H. Prognostic value of HMGA2, P16, and HPV in oral squamous cell carcinomas. J. Craniomaxillofac. Surg. 2016, 44, 1422–1429. [Google Scholar] [CrossRef]
- Chang, K.P.; Lin, S.J.; Liu, S.C.; Yi, J.S.; Chien, K.Y.; Chi, L.M.; Kao, H.K.; Liang, Y.; Lin, Y.T.; Chang, Y.S.; et al. Low-molecular-mass secretome profiling identifies HMGA2 and MIF as prognostic biomarkers for oral cavity squamous cell carcinoma. Sci. Rep. 2015, 5, 11689. [Google Scholar] [CrossRef] [Green Version]
- Fang, C.Y.; Liew, P.L.; Chen, C.L.; Lin, Y.H.; Fang, C.L.; Chen, W.Y. High HMGA2 expression correlates with reduced recurrence-free survival and poor overall survival in oral squamous cell carcinoma. Anticancer Res. 2017, 37, 1891–1899. [Google Scholar] [PubMed]
- Sterenczak, K.A.; Eckardt, A.; Kampmann, A.; Willenbrock, S.; Eberle, N.; Langer, F.; Kleinschmidt, S.; Hewicker-Trautwein, M.; Kreipe, H.; Nolte, I.; et al. HMGA1 and HMGA2 expression and comparative analyses of HMGA2, Lin28 and let-7 miRNAs in oral squamous cell carcinoma. BMC Cancer 2014, 14, 694. [Google Scholar] [CrossRef]
- Wang, X.; Liu, X.; Li, A.Y.; Chen, L.; Lai, L.; Lin, H.H.; Hu, S.; Yao, L.; Peng, J.; Loera, S.; et al. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin. Cancer Res. 2011, 17, 2570–2580. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Y.; Xu, X.; Cao, H.; Sahengbieke, S.; Sheng, H.; Huang, Q.; Lai, M. Transcriptional activation of FN1 and IL11 by HMGA2 promotes the malignant behavior of colorectal cancer. Carcinogenesis 2016, 37, 511–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiger, T.R.; Peeper, D.S. Metastasis mechanisms. Biochim. Biophys. Acta 2009, 1796, 293–308. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Muthusamy, B.P.; Saeteurn, K.Y. Signaling pathway cooperation in TGF-beta-induced epithelial-mesenchymal transition. Curr. Opin. Cell Biol. 2014, 31, 56–66. [Google Scholar] [CrossRef]
- Chen, S.S.; Tang, C.H.; Chie, M.J.; Tsai, C.H.; Fong, Y.C.; Lu, Y.C.; Chen, W.C.; Lai, C.T.; Wei, C.Y.; Tai, H.C.; et al. Resistin facilitates VEGF-A-dependent angiogenesis by inhibiting miR-16-5p in human chondrosarcoma cells. Cell Death Dis. 2019, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Hasina, R.; Lingen, M.W. Angiogenesis in oral cancer. J. Dent. Educ. 2001, 65, 1282–1290. [Google Scholar]
- Baeriswyl, V.; Christofori, G. The angiogenic switch in carcinogenesis. Semin. Cancer Biol. 2009, 19, 329–337. [Google Scholar] [CrossRef]
- Helmke, B.M.; Markowski, D.N.; Muller, M.H.; Sommer, A.; Muller, J.; Moller, C.; Bullerdiek, J. HMGA proteins regulate the expression of FGF2 in uterine fibroids. Mol. Hum. Reprod. 2011, 17, 135–142. [Google Scholar] [CrossRef]
- Mariz, B.A.L.A.; Soares, C.D.; de Carvalho, M.G.F.; Jorge-Junior, J. FGF-2 and FGFR-1 might be independent prognostic factors in oral tongue squamous cell carcinoma. Histopathology 2019, 74, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.J.; Kahata, K.; Idas, O.; Thuault, S.; Heldin, C.H.; Moustakas, A. The high mobility group A2 protein epigenetically silences the Cdh1 gene during epithelial-to-mesenchymal transition. Nucleic Acids Res. 2015, 43, 162–178. [Google Scholar] [CrossRef] [PubMed]







| Characteristics | Total | HMGA2 Status | p-Value | |
|---|---|---|---|---|
| High Expression | Low Expression | |||
| n (%) | n (%) | |||
| 110 | 42 (38.2) | 68 (61.8) | ||
| Age, years | ||||
| Median | 67 | 65.9 | 67.7 | |
| Range | 30–87 | 30–85 | 33–87 | |
| ≤65 | 45 | 19 (42.2) | 26 (57.8) | 0.468 |
| >65 | 65 | 23 (35.4) | 42 (64.6) | |
| Sex | ||||
| Male | 66 | 28 (42.4) | 38 (57.6) | 0.262 |
| Female | 44 | 14 (31.2) | 30 (68.2) | |
| Primary site | ||||
| Tongue | 34 | 13 (38.2) | 21 (61.8) | 0.532 |
| Mandible gingiva | 25 | 12 (48.0) | 13 (52.0) | |
| Maxilla gingiva | 21 | 8 (38.1) | 13 (61.9) | |
| Buccal mucosa | 16 | 4 (25.0) | 12 (75.0) | |
| Oral floor | 13 | 4 (30.8) | 9 (69.2) | |
| Palate | 1 | 1 (100) | 0 (0) | |
| T-stage | ||||
| T1, T2 | 37 | 11 (29.7) | 26 (70.3) | 0.08 |
| T3 | 25 | 7 (28.0) | 18 (72.0) | |
| T4 | 48 | 24 (50.0) | 24 (50.0) | |
| N-stage | ||||
| N0 | 34 | 7 (20.6) | 27 (79.4) | 0.011 * |
| ≥N1 | 76 | 35 (46.1) | 41 (53.9) | |
| Clinical stage | ||||
| II | 13 | 2 (15.4) | 11 (84.6) | 0.142 |
| III | 26 | 9 (34.6) | 17 (65.4) | |
| IV | 71 | 31 (43.7) | 40 (56.3) | |
| Differentiation | ||||
| Well | 90 | 35 (38.9) | 55 (61.1) | 0.746 |
| Moderate | 20 | 7 (35.0) | 13 (65.0) | |
| Local recurrence | ||||
| Yes | 28 | 11 (39.3) | 17 (60.7) | 0.889 |
| No | 82 | 31 (37.8) | 51 (62.2) | |
| Distant metastasis | ||||
| Yes | 16 | 13 (81.3) | 3 (18.7) | <0.001 ** |
| No | 94 | 29 (30.9) | 65 (69.1) | |
| Characteristics | Assigned Score | OS | DFS | ||
|---|---|---|---|---|---|
| Hazard Ratio (95% CI) | p-Value | Hazard Ratio (95% CI) | p-Value | ||
| HMGA2 expression | |||||
| Low | 0 | 1.254 (0.528–3.012) | 0.609 | 1.195 (0.545–2.650) | 0.657 |
| High | 1 | ||||
| N-stage | |||||
| N0 | 0 | 1.290 (0.508–3.692) | 0.603 | 1.068 (0.455–2.639) | 0.882 |
| ≥N1 | 1 | ||||
| Local recurrence | |||||
| No | 0 | 10.33 (4.640–24.72) | <0.001 ** | 14.09 (6.553–32.68) | <0.001 ** |
| Yes | 1 | ||||
| Distant metastasis | |||||
| No | 0 | 27.89 (10.09–80.16) | <0.001 ** | 5.222 (2.247–12.19) | <0.001 ** |
| Yes | 1 | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakata, J.; Hirosue, A.; Yoshida, R.; Kawahara, K.; Matsuoka, Y.; Yamamoto, T.; Nakamoto, M.; Hirayama, M.; Takahashi, N.; Nakamura, T.; et al. HMGA2 Contributes to Distant Metastasis and Poor Prognosis by Promoting Angiogenesis in Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2019, 20, 2473. https://doi.org/10.3390/ijms20102473
Sakata J, Hirosue A, Yoshida R, Kawahara K, Matsuoka Y, Yamamoto T, Nakamoto M, Hirayama M, Takahashi N, Nakamura T, et al. HMGA2 Contributes to Distant Metastasis and Poor Prognosis by Promoting Angiogenesis in Oral Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2019; 20(10):2473. https://doi.org/10.3390/ijms20102473
Chicago/Turabian StyleSakata, Junki, Akiyuki Hirosue, Ryoji Yoshida, Kenta Kawahara, Yuichiro Matsuoka, Tatsuro Yamamoto, Masafumi Nakamoto, Masatoshi Hirayama, Nozomu Takahashi, Takuya Nakamura, and et al. 2019. "HMGA2 Contributes to Distant Metastasis and Poor Prognosis by Promoting Angiogenesis in Oral Squamous Cell Carcinoma" International Journal of Molecular Sciences 20, no. 10: 2473. https://doi.org/10.3390/ijms20102473
APA StyleSakata, J., Hirosue, A., Yoshida, R., Kawahara, K., Matsuoka, Y., Yamamoto, T., Nakamoto, M., Hirayama, M., Takahashi, N., Nakamura, T., Arita, H., Nakashima, H., Nagata, M., Hiraki, A., Shinohara, M., & Nakayama, H. (2019). HMGA2 Contributes to Distant Metastasis and Poor Prognosis by Promoting Angiogenesis in Oral Squamous Cell Carcinoma. International Journal of Molecular Sciences, 20(10), 2473. https://doi.org/10.3390/ijms20102473