Effects of Abscisic Acid and Salicylic Acid on Gene Expression in the Antiviral RNA Silencing Pathway in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. Binding Sites of Several TFs Regulated by ABA and SA Are Located in the Promoter Regions of Genes Belonging to the AGO, DCL, and RDR Families
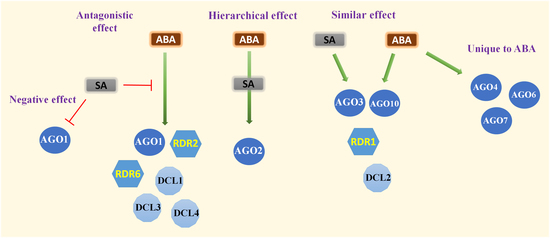
2.2. ABA and SA Exert Similar Effects on Expression of Several Genes in the RNA Silencing Pathway
2.3. Susceptibility to BaMV Is Not Greater in the ABA/SA Double Mutant Than in the Single Mutants
3. Discussion
4. Materials and Methods
4.1. Coexpression and Promoter Analyses
4.2. Plant Materials
4.3. Hormone Treatments
5.4. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
4.5. Protein Analyses
5.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef]
- Ton, J.; Flors, V.; Mauch-Mani, B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef]
- Alazem, M.; He, M.H.; Moffett, P.; Lin, N.S. Abscisic Acid Induces Resistance against Bamboo Mosaic Virus through Argonaute2 and 3. Plant Physiol. 2017, 174, 339–355. [Google Scholar] [CrossRef] [PubMed]
- Klessig, D.F.; Choi, H.W.; Dempsey, D.A. Systemic Acquired Resistance and Salicylic Acid: Past, Present, and Future. Mol. Plant Microbe Interact 2018, MPMI03180067CR. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, M.; Ishikawa, A.; Jikumaru, Y.; Seki, M.; Umezawa, T.; Asami, T.; Maruyama-Nakashita, A.; Kudo, T.; Shinozaki, K.; Yoshida, S.; Nakashita, H. Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 2008, 20, 1678–1692. [Google Scholar] [CrossRef]
- Fan, J.; Hill, L.; Crooks, C.; Doerner, P.; Lamb, C. Abscisic Acid Has a Key Role in Modulating Diverse Plant-Pathogen Interactions. Plant Physiol. 2009, 150, 1750–1761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zabala, M.D.; Bennett, M.H.; Truman, W.H.; Grant, M.R. Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 2009, 59, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Denance, N.; Sanchez-Vallet, A.; Goffner, D.; Molina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef]
- Prodhan, M.Y.; Munemasa, S.; Nahar, M.N.E.N.; Nakamura, Y.; Murata, Y. Guard Cell Salicylic Acid Signaling Is Integrated into Abscisic Acid Signaling via the Ca2+/CPK-Dependent Pathway. Plant Physiol. 2018, 178, 441–450. [Google Scholar] [CrossRef]
- Alazem, M.; Lin, N.S. Roles of plant hormones in the regulation of host-virus interactions. Mol. Plant Pathol. 2015, 16, 529–540. [Google Scholar] [CrossRef]
- Alazem, M.; Lin, N.S. Antiviral Roles of Abscisic Acid in Plants. Front. Plant Sci. 2017, 8, 1760. [Google Scholar] [CrossRef]
- Alazem, M.; Tseng, K.C.; Chang, W.C.; Seo, J.K.; Kim, K.H. Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus. Viruses 2018, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Alamillo, J.M.; Saenz, P.; Garcia, J.A. Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. Plant J. 2006, 48, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cui, X.; Meng, Z.; Huang, X.; Xie, Q.; Wu, H.; Jin, H.; Zhang, D.; Liang, W. Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 2012, 158, 1279–1292. [Google Scholar] [CrossRef] [PubMed]
- Jaubert, M.; Bhattacharjee, S.; Mello, A.F.; Perry, K.L.; Moffett, P. ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 2011, 156, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Hunter, L.J.; Westwood, J.H.; Heath, G.; Macaulay, K.; Smith, A.G.; Macfarlane, S.A.; Palukaitis, P.; Carr, J.P. Regulation of RNA-dependent RNA polymerase 1 and isochorismate synthase gene expression in Arabidopsis. PLoS ONE 2013, 8, e66530. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Fu, S.F.; Li, Z.; Murphy, A.M.; Dobson, E.A.; Garland, L.; Chaluvadi, S.R.; Lewsey, M.G.; Nelson, R.S.; Carr, J.P. Salicylic acid treatment and expression of an RNA-dependent RNA polymerase 1 transgene inhibit lethal symptoms and meristem invasion during tobacco mosaic virus infection in Nicotiana benthamiana. BMC Plant Biol. 2016, 16, 15. [Google Scholar] [CrossRef]
- Lee, C.H.; Carroll, B.J. Evolution and Diversification of Small RNA Pathways in Flowering Plants. Plant Cell Physiol. 2018, 59, 2169–2187. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Kontra, L.; Burgyan, J. viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef]
- Chow, C.N.; Zheng, H.Q.; Wu, N.Y.; Chien, C.H.; Huang, H.D.; Lee, T.Y.; Chiang-Hsieh, Y.F.; Hou, P.F.; Yang, T.Y.; Chang, W.C. PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016, 44, D1154–D1160. [Google Scholar] [CrossRef]
- Hsu, Y.H.; Tsai, C.H.; Lin, N.S. Editorial: Molecular Biology of Bamboo mosaic Virus-A Type Member of the Potexvirus Genus. Front. Microbiol. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.S.; Lin, B.Y.; Lo, N.W.; Hu, C.C.; Chow, T.Y.; Hsu, Y.H. Nucleotide sequence of the genomic RNA of bamboo mosaic potexvirus. J. Gen. Virol. 1994, 75, 2513–2518. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.S.; Lin, F.Z.; Huang, T.Y.; Hsu, Y.H. Genome Properties of Bamboo Mosaic-Virus. Phytopathology 1992, 82, 731–734. [Google Scholar] [CrossRef]
- DiMaio, F.; Chen, C.C.; Yu, X.; Frenz, B.; Hsu, Y.H.; Lin, N.S.; Egelman, E.H. The molecular basis for flexibility in the flexible filamentous plant viruses. Nat. Struct. Mol. Biol. 2015, 22, 642–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.Q.; Wu, N.Y.; Chow, C.N.; Tseng, K.C.; Chien, C.H.; Hung, Y.C.; Li, G.Z.; Chang, W.C. EXPath tool-a system for comprehensively analyzing regulatory pathways and coexpression networks from high-throughput transcriptome data. DNA Res. 2017, 24, 371–375. [Google Scholar] [CrossRef]
- Jensen, M.K.; Hagedorn, P.H.; de Torres-Zabala, M.; Grant, M.R.; Rung, J.H.; Collinge, D.B.; Lyngkjaer, M.F. Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 2008, 56, 867–880. [Google Scholar] [CrossRef]
- Wu, Y.; Deng, Z.; Lai, J.; Zhang, Y.; Yang, C.; Yin, B.; Zhao, Q.; Zhang, L.; Li, Y.; Yang, C.; Xie, Q. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009, 19, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hua, D.; He, J.; Duan, Y.; Chen, Z.; Hong, X.; Gong, Z. Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 2011, 7, e1002172. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Fujita, Y.; Katsura, K.; Maruyama, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 2006, 60, 51–68. [Google Scholar] [CrossRef]
- Delmas, F.; Sankaranarayanan, S.; Deb, S.; Widdup, E.; Bournonville, C.; Bollier, N.; Northey, J.G.B.; McCourt, P.; Samuel, M.A. ABI3 controls embryo degreening through Mendel’s I locus. Proc. Natl. Acad. Sci. USA 2013, 110, E3888–E3894. [Google Scholar] [CrossRef]
- Acevedo-Hernandez, G.J.; Leon, P.; Herrera-Estrella, L.R. Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J. 2005, 43, 506–519. [Google Scholar] [CrossRef]
- Kaliff, M.; Staal, J.; Myrenas, M.; Dixelius, C. ABA is required for Leptosphaeria maculans resistance via ABI1- and ABI4-dependent signaling. Mol. Plant Microbe Ind. 2007, 20, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Shkolnik-Inbar, D.; Bar-Zvi, D. ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 2010, 22, 3560–3573. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Zhang, H.; Wang, S.; Chen, M.; Wu, Y.; Tang, S.; Liu, C.; Feng, Y.; Cao, X.; Xie, Q. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. PLoS Genet. 2013, 9, e1003577. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.Z.; Chen, Y.; Wang, C.; Kong, Y.H.; Wu, W.H.; Chen, Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014, 80, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Molina, L.; Mongrand, S.; Chua, N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the AB15 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 2001, 98, 4782–4787. [Google Scholar] [CrossRef]
- Carles, C.; Bies-Etheve, N.; Aspart, L.; Leon-Kloosterziel, K.M.; Koornneef, M.; Echeverria, M.; Delseny, M. Regulation of Arabidopsis thaliana Em genes: Role of ABI5. Plant J. 2002, 30, 373–383. [Google Scholar] [CrossRef]
- Bensmihen, S.; Rippa, S.; Lambert, G.; Jublot, D.; Pautot, V.; Granier, F.; Giraudat, J.; Parcy, F. The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 2002, 14, 1391–1403. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002, 32, 317–328. [Google Scholar] [CrossRef] [Green Version]
- Finkelstein, R.; Gampala, S.S.; Lynch, T.J.; Thomas, T.L.; Rock, C.D. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 2005, 59, 253–267. [Google Scholar] [CrossRef]
- Kim, M.J.; Park, M.J.; Seo, P.J.; Song, J.S.; Kim, H.J.; Park, C.M. Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem. J. 2012, 448, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Kim, M.J.; Park, J.Y.; Kim, S.Y.; Jeon, J.; Lee, Y.H.; Kim, J.; Park, C.M. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010, 61, 661–671. [Google Scholar] [CrossRef]
- Lechner, E.; Leonhardt, N.; Eisler, H.; Parmentier, Y.; Alioua, M.; Jacquet, H.; Leung, J.; Genschik, P. MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev. Cell 2011, 21, 1116–1128. [Google Scholar] [CrossRef]
- Johannesson, H.; Wang, Y.; Hanson, J.; Engstrom, P. The Arabidopsis thaliana homeobox gene ATHB5 is a potential regulator of abscisic acid responsiveness in developing seedlings. Plant Mol. Biol. 2003, 51, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Yu, D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009, 9, 96. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Lee, S.J.; Kang, J.Y.; Park, H.J.; Kim, M.D.; Bae, M.S.; Choi, H.I.; Kim, S.Y. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 2010, 153, 716–727. [Google Scholar] [CrossRef]
- Kim, J.S.; Mizoi, J.; Yoshida, T.; Fujita, Y.; Nakajima, J.; Ohori, T.; Todaka, D.; Nakashima, K.; Hirayama, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. An ABRE Promoter Sequence is Involved in Osmotic Stress-Responsive Expression of the DREB2A Gene, Which Encodes a Transcription Factor Regulating Drought-Inducible Genes in Arabidopsis. Plant Cell Physiol. 2011, 52, 2136–2146. [Google Scholar] [CrossRef]
- Tang, W.; Ji, Q.; Huang, Y.; Jiang, Z.; Bao, M.; Wang, H.; Lin, R. FAR-RED ELONGATED HYPOCOTYL3 and FAR-RED IMPAIRED RESPONSE1 transcription factors integrate light and abscisic acid signaling in Arabidopsis. Plant Physiol. 2013, 163, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Van Hengel, A.J.; Barber, C.; Roberts, K. The expression patterns of arabinogalactan-protein AtAGP30 and GLABRA2 reveal a role for abscisic acid in the early stages of root epidermal patterning. Plant J. 2004, 39, 70–83. [Google Scholar] [CrossRef]
- Soderman, E.; Mattsson, J.; Engstrom, P. The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 1996, 10, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.E.; Overnas, E.; Johansson, H.; Rada-Iglesias, A.; Engstrom, P. The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 2012, 80, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Yamamoto, A.; Kagaya, Y.; Toyoshima, R.; Kagaya, M.; Takeda, S.; Hattori, T. Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 2009, 58, 843–856. [Google Scholar] [CrossRef]
- Brocard-Gifford, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol. 2003, 131, 78–92. [Google Scholar] [CrossRef]
- Reyes, J.L.; Chua, N.H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007, 49, 592–606. [Google Scholar] [CrossRef]
- Tran, L.S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Yanhui, C.; Xiaoyuan, Y.; Kun, H.; Meihua, L.; Jigang, L.; Zhaofeng, G.; Zhiqiang, L.; Yunfei, Z.; Xiaoxiao, W.; Xiaoming, Q.; Yunping, S.; Li, Z.; Xiaohui, D.; Jingchu, L.; Xing-Wang, D.; Zhangliang, C.; Hongya, G.; Li-Jia, Q. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, X.; Guo, L.; Lu, F.; Feng, X.; He, K.; Wei, L.; Chen, Z.; Qu, L.J.; Gu, H. A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. J. Exp. Bot. 2006, 57, 1263–1273. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.G.; Singh, K.B. Characterization of salicylic acid-responsive, arabidopsis Dof domain proteins: Overexpression of OBP3 leads to growth defects. Plant J. 2000, 21, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.G.; Foley, R.C.; Onate-Sanchez, L.; Lin, C.; Singh, K.B. Target genes for OBP3, a Dof transcription factor, include novel basic helix-loop-helix domain proteins inducible by salicylic acid. Plant J. 2003, 35, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Boden, E.; Arias, J. Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 2003, 15, 1846–1858. [Google Scholar] [CrossRef] [PubMed]
- Zander, M.; La Camera, S.; Lamotte, O.; Metraux, J.P.; Gatz, C. Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 2010, 61, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, J.P.; Menossi, M.; Thibaud-Nissen, F.; Town, C.D. Functional analysis of a TGA factor-binding site located in the promoter region controlling salicylic acid-induced NIMIN-1 expression in Arabidopsis. Genet. Mol Res. 2010, 9, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Thibaud-Nissen, F.; Wu, H.; Richmond, T.; Redman, J.C.; Johnson, C.; Green, R.; Arias, J.; Town, C.D. Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J. 2006, 47, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Huh, S.U.; Kojima, M.; Sakakibara, H.; Paek, K.H.; Hwang, I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 2010, 19, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.M.; Trifa, Y.; Silva, H.; Pontier, D.; Lam, E.; Shah, J.; Klessig, D.F. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Ind. 2000, 13, 191–202. [Google Scholar] [CrossRef]
- Kang, H.G.; Klessig, D.F. Salicylic acid-inducible Arabidopsis CK2-like activity phosphorylates TGA2. Plant Mol. Biol. 2005, 57, 541–557. [Google Scholar] [CrossRef]
- Duan, M.R.; Nan, J.; Liang, Y.H.; Mao, P.; Lu, L.; Li, L.; Wei, C.; Lai, L.; Li, Y.; Su, X.D. DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res. 2007, 35, 1145–1154. [Google Scholar] [CrossRef] [Green Version]
- Robatzek, S.; Somssich, I.E. A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 2001, 28, 123–133. [Google Scholar] [CrossRef]
- Yu, D.; Chen, C.; Chen, Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 2001, 13, 1527–1540. [Google Scholar] [CrossRef]
- Lai, Z.; Vinod, K.; Zheng, Z.; Fan, B.; Chen, Z. Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 2008, 8, 68. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, L.; Yu, D. Wounding-induced WRKY8 is involved in basal defense in Arabidopsis. Mol. Plant Microbe Interact 2010, 23, 558–565. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, L.; Wang, H.; Zhang, L.; Wang, F.; Yu, D. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 2013, 74, 730–745. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, L.; Li, D.; Wang, F.; Yu, D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, E1963–E1971. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Q.; Li, Z.; Staswick, P.E.; Wang, M.; Zhu, Y.; He, Z. Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 2007, 145, 450–464. [Google Scholar] [CrossRef]
- Scarpeci, T.E.; Zanor, M.I.; Mueller-Roeber, B.; Valle, E.M. Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol. Biol. 2013, 83, 265–277. [Google Scholar] [CrossRef]
- Li, J.; Besseau, S.; Toronen, P.; Sipari, N.; Kollist, H.; Holm, L.; Palva, E.T. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013, 200, 457–472. [Google Scholar] [CrossRef]
- Wu, H.Y.; Liu, K.H.; Wang, Y.C.; Wu, J.F.; Chiu, W.L.; Chen, C.Y.; Wu, S.H.; Sheen, J.; Lai, E.M. AGROBEST: An efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods 2014, 10, 19. [Google Scholar] [CrossRef]
- Song, L.; Huang, S.C.; Wise, A.; Castanon, R.; Nery, J.R.; Chen, H.; Watanabe, M.; Thomas, J.; Bar-Joseph, Z.; Ecker, J.R. A transcription factor hierarchy defines an environmental stress response network. Science 2016, 354, 6312. [Google Scholar] [CrossRef]
- Sun, D.; Nandety, R.S.; Zhang, Y.; Reid, M.S.; Niu, L.; Jiang, C.Z. A petunia ethylene-responsive element binding factor, PhERF2, plays an important role in antiviral RNA silencing. J. Exp. Bot. 2016, 67, 3353–3365. [Google Scholar] [CrossRef]
- Yoon, E.K.; Yang, J.H.; Lim, J.; Kim, S.H.; Kim, S.K.; Lee, W.S. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 2010, 38, 1382–1391. [Google Scholar] [CrossRef]
- Lewsey, M.G.; Murphy, A.M.; Maclean, D.; Dalchau, N.; Westwood, J.H.; Macaulay, K.; Bennett, M.H.; Moulin, M.; Hanke, D.E.; Powell, G.; Smith, A.G.; Carr, J.P. Disruption of two defensive signaling pathways by a viral RNA silencing suppressor. Mol. Plant Microbe Interact 2010, 23, 835–845. [Google Scholar] [CrossRef]
- Morris, K.; Mackerness, S.A.H.; Page, T.; John, C.F.; Murphy, A.M.; Carr, J.P.; Buchanan-Wollaston, V. Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 2000, 23, 677–685. [Google Scholar] [CrossRef]
- Qin, J.; Ma, X.; Yi, Z.; Tang, Z.; Meng, Y. A transcriptome-wide study on the microRNA- and the Argonaute 1-enriched small RNA-mediated regulatory networks involved in plant leaf senescence. Plant Biol. 2016, 18, 197–205. [Google Scholar] [CrossRef]
- De Vleesschauwer, D.; Xu, J.; Hofte, M. Making sense of hormone-mediated defense networking: From rice to Arabidopsis. Front. Plant Sci. 2014, 5, 611. [Google Scholar] [CrossRef]
- Lee, S.; Park, C.M. Modulation of reactive oxygen species by salicylic acid in Arabidopsis seed germination under high salinity. Plant Signal. Behav. 2010, 5, 1534–1536. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, J.; Li, S.; Kakan, X.; Zhou, Y.; Miao, Y.; Wang, F.; Qin, H.; Huang, R. Ascorbic Acid Integrates the Antagonistic Modulation of Ethylene and Abscisic Acid in the Accumulation of Reactive Oxygen Species. Plant Physiol. 2019, 179, 1861–1875. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Alazem, M.; Lin, K.Y.; Lin, N.S. The abscisic acid pathway has multifaceted effects on the accumulation of Bamboo mosaic virus. Mol. Plant Microbe Interact 2014, 27, 177–189. [Google Scholar] [CrossRef]
- Prasanth, K.R.; Huang, Y.W.; Liou, M.R.; Wang, R.Y.; Hu, C.C.; Tsai, C.H.; Meng, M.; Lin, N.S.; Hsu, Y.H. Glyceraldehyde 3-phosphate dehydrogenase negatively regulates the replication of Bamboo mosaic virus and its associated satellite RNA. J. Virol. 2011, 85, 8829–8840. [Google Scholar] [CrossRef]
- Despres, C.; Chubak, C.; Rochon, A.; Clark, R.; Bethune, T.; Desveaux, D.; Fobert, P.R. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 2003, 15, 2181–2191. [Google Scholar] [CrossRef]
- Katagiri, F.; Lam, E.; Chua, N.H. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 1989, 340, 727–730. [Google Scholar] [CrossRef]
- Xiang, C.; Miao, Z.; Lam, E. DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol. Biol. 1997, 34, 403–415. [Google Scholar] [CrossRef]
- Klinedinst, S.; Pascuzzi, P.; Redman, J.; Desai, M.; Arias, J. A xenobiotic-stress-activated transcription factor and its cognate target genes are preferentially expressed in root tip meristems. Plant Mol. Biol. 2000, 42, 679–688. [Google Scholar] [CrossRef]



| TF | Acc. No. | Promoters of | TF Family | Regulation | GO/Functional Description | Reference | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AGO | RDR | DCL | |||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 9 | 10 | 1 | 2 | 6 | 1 | 2 | 3 | 4 | ||||||
| ABI3 | AT3G24650 | ● | ● | ● | ● | B3 | ABA | ABA-activated signalling pathway - response to ABA | [29,30] | ||||||||||||
| ABI4 | AT2G40220 | ● | ERF | ABA | ABA-activated signalling pathway | [31,32,33,34,35] | |||||||||||||||
| ABI5 | AT2G36270 | ● | ● | bZIP | ABA | ABA-activated signalling pathway - response to ABA | [29,36,37,38,39,40] | ||||||||||||||
| ANAC062 | AT3G49530 | ● | NAC | ABA | Plays a regulatory role in ABA-mediated drought-resistance. Mediates induction of pathogenesis-related (PR) genes independently of salicylic signalling in response to cold | [41,42] | |||||||||||||||
| ATAF1 | AT1G01720 | ● | ● | ● | NAC | ABA | Negative regulation of ABA-activated signalling pathway | [26,27] | |||||||||||||
| AtHB33 | AT1G75240 | ● | ● | ● | ● | ● | ● | ZF-HD | ABA | Repressed by ABA and ARF2, Regulators in the ABA signal pathway that confers sensitivity to ABA in an ARF2-dependent manner. | [27] | ||||||||||
| ATHB5 | AT5G65310 | ● | HD-ZIP | ABA | Probable transcription factor that acts as a positive regulator of ABA-responsiveness, mediating the inhibitory effect of ABA on growth during seedling establishment. Binds to the DNA sequence 5′-CAATNATTG-3′. | [27] | |||||||||||||||
| AT-HSFA9 | AT5G54070 | ● | HSF | ABA | A member of Heat Stress Transcription Factor (Hsf) family. Not responding to heat stress. Is regulated by the seed-specific transcription factor ABI3. In turn, it regulates other heat stress proteins including Hsp17.4-CI, Hsp17.7-CII and Hsp101 during seed maturation. | [44] | |||||||||||||||
| DREB19 | AT2G38340 | ● | ● | ● | ● | ERF | ABA | Induced by ABA treatment. Transcriptional activator that binds specifically to the DNA sequence 5′-[AG]CCGAC-3′. Binding to the C-repeat/DRE element mediates ABA-inducible transcription | [46] | ||||||||||||
| DREB2 | AT5G05410 | ● | ● | ● | ● | ● | ● | ● | ERF | ABA | the ABA-dependent pathway plays a positive role in the osmotic stress-responsive expression of DREB2A | [47,48] | |||||||||
| FHY3 | AT3G22170 | ● | FAR1 | ABA | FHY3 and FAR1 are positive regulators of ABA signalling and provide insight into the integration of light and ABA signalling | [49] | |||||||||||||||
| GL2 | AT1G79840 | ● | HD-ZIP | ABA | The expression patterns of arabinogalactan-protein AtAGP30 and GLABRA2 reveal a role for ABA in the early stages of root epidermal patterning. | [50] | |||||||||||||||
| HB-7 | AT5G46880 | ● | ● | HD-ZIP | ABA | NDUCTION: By water deficit, by ABA and by salt stress | [51,52] | ||||||||||||||
| ICE1 | AT3G26744 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | bHLH | ABA | INDUCTION: By high-salt stress, cold stress and ABA treatment. | [53] | ||||||
| LEC1 | AT1G21970 | ● | ● | NF-YB | ABA | ABA-activated signalling pathway | [55,57] | ||||||||||||||
| MYB33 | AT5G06100 | ● | ● | ● | MYB | ABA | positive regulation of ABA-activated signalling pathway | [56] | |||||||||||||
| NAC3 | AT3G15500 | ● | NAC | ABA | Strongly induced by high salinity. Slightly up-regulated by drought, ABA and jasmonic acid. Not induced by cold treatment. | [57] | |||||||||||||||
| WRKY2 | AT5G56270 | ● | WRKY | ABA | Transcription factor. Regulates WOX8 and WOX9 expression and basal cell division patterns during early embryogenesis. Interacts specifically with the W box (5′-(T)TGAC[CT]-3′), a frequently occurring elicitor-responsive cis-acting element. Required to repolarize the zygote from a transient symmetric state | [45] | |||||||||||||||
| ATHB16 | AT4G40060 | ● | HD-ZIP | ABA | MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate ABA signalling. | [43] | |||||||||||||||
| MYB59 | AT5G59780 | ● | MYB | SA | Isoform MYB59-1 is induced by JA, SA, gibberellic acid, and ethylene | [58,59] | |||||||||||||||
| MYB65 | AT3G11440 | ● | MYB | SA | response to SA | [58] | |||||||||||||||
| OBP1 | AT3G50410 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | bZIP | SA | Induced by SA, Constitutively expressed in the whole plant | [59] | ||||||
| TGA3 | AT1G22070 | ● | bZIP | SA | systemic acquired resistance, SA mediated signalling pathway | [62,65,66,67,68] | |||||||||||||||
| TGA4 | AT5G10030 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | bZIP | SA | Binding to the as-1-like cis elements mediate auxin- and SA-inducible transcription. May be involved in the induction of the systemic acquired resistance (SAR) via its interaction with NPR1. | [60,67] | |||||
| WRKY1 | AT2G04880 | ● | ● | ● | ● | ● | WRKY | SA | SA-mediated signalling pathway | [69] | |||||||||||
| WRKY15 | AT2G23320 | ● | WRKY | SA | Induced by SA | [71] | |||||||||||||||
| WRKY21 | AT2G30590 | ● | ● | ● | WRKY | SA | Induced by SA | [71] | |||||||||||||
| WRKY26 | AT5G07100 | ● | WRKY | SA | Induced by SA | [71,76] | |||||||||||||||
| WRKY3 | AT2G03340 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | WRKY | SA | induced by SA and during leaf senescence | [70,71] | ||||||
| WRKY30 | AT5G24110 | ● | WRKY | SA | response to SA | [77] | |||||||||||||||
| WRKY4 | AT1G13960 | ● | ● | ● | ● | ● | ● | WRKY | SA | INDUCTION: By biotic and abiotic stresses such as pathogen infection, SA, JA, ACC | [72] | ||||||||||
| WRKY54 | AT2G40750 | ● | WRKY | SA | WRKY70 and WRKY54 co-operate as negative regulators of stomatal closure and, consequently, osmotic stress tolerance in Arabidopsis, suggesting that they have an important role, not only in plant defence, but also in abiotic stress signalling. WRKY70 and WRKY54 are positive regulators of plant defence, and co-operate as negative regulators of SA biosynthesis and senescence. | [78] | |||||||||||||||
| OBP3 | AT3G55370 | ● | bZIP | SA- JA | Induced by SA, Repressed by JA | [60,61] | |||||||||||||||
| TGA2 | AT5G06950 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | bZIP | SA- JA/Et | Required to induce the systemic acquired resistance (SAR) via the regulation of pathogenesis-related genes expression | [62,63,64,65] | |||||
| TGA5 | AT5G06960 | ● | ● | ● | ● | ● | ● | ● | ● | ● | bZIP | SA- JA/Et | May be involved in the induction of the systemic acquired resistance (SAR) via its interaction with NPR1. | [63,67,68] | |||||||
| TGA6 | AT3G12250 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | bZIP | SA- JA/Et | May be involved in the induction of the systemic acquired resistance (SAR) via its interaction with NPR1. | [63,67,68] | |||||
| WRKY8 | AT5G46350 | ● | WRKY | SA/ABA | Induced by wounding, ABA, SA, H2O2 and infection with P.syringae pv. tomato DC3000 and B.cinerea | [73,74,76] | |||||||||||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alazem, M.; Kim, K.-H.; Lin, N.-S. Effects of Abscisic Acid and Salicylic Acid on Gene Expression in the Antiviral RNA Silencing Pathway in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2538. https://doi.org/10.3390/ijms20102538
Alazem M, Kim K-H, Lin N-S. Effects of Abscisic Acid and Salicylic Acid on Gene Expression in the Antiviral RNA Silencing Pathway in Arabidopsis. International Journal of Molecular Sciences. 2019; 20(10):2538. https://doi.org/10.3390/ijms20102538
Chicago/Turabian StyleAlazem, Mazen, Kook-Hyung Kim, and Na-Sheng Lin. 2019. "Effects of Abscisic Acid and Salicylic Acid on Gene Expression in the Antiviral RNA Silencing Pathway in Arabidopsis" International Journal of Molecular Sciences 20, no. 10: 2538. https://doi.org/10.3390/ijms20102538
APA StyleAlazem, M., Kim, K. -H., & Lin, N. -S. (2019). Effects of Abscisic Acid and Salicylic Acid on Gene Expression in the Antiviral RNA Silencing Pathway in Arabidopsis. International Journal of Molecular Sciences, 20(10), 2538. https://doi.org/10.3390/ijms20102538