Organization of DNA in Mammalian Mitochondria
Abstract
:1. DNA Compaction
2. The Mitochondrial Nucleoid
3. mtDNA Compaction in Yeast
4. mtDNA Compaction in Mammals
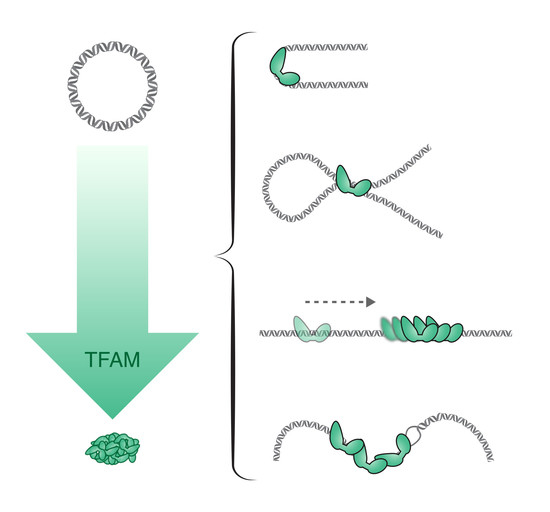
5. The Regulatory Role of TFAM
5.1. mtDNA Replication and Transcription
5.2. The Role of TFAM in the Regulation of mtDNA Replication and Transcription
Funding
Acknowledgments
Conflicts of Interest
References
- Van Noort, V.; Snel, B.; Huynen, M.A. The yeast coexpression network has a small-world, scale-free architecture and can be explained by a simple model. EMBO Rep. 2004, 5, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Dame, R.T.; Noom, M.C.; Wuite, G.J. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature 2006, 444, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Dillon, S.C.; Dorman, C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010, 8, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Dorman, C.J. H-NS-like nucleoid-associated proteins, mobile genetic elements and horizontal gene transfer in bacteria. Plasmid 2014, 75, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browning, D.F.; Grainger, D.C.; Busby, S.J. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 2010, 13, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Krogh, T.J.; Moller-Jensen, J.; Kaleta, C. Impact of chromosomal architecture on the function and evolution of bacterial genomes. Front. Microbiol. 2018, 9, 2019. [Google Scholar] [CrossRef]
- Skoko, D.; Yan, J.; Johnson, R.C.; Marko, J.F. Low-force DNA condensation and discontinuous high-force decondensation reveal a loop-stabilizing function of the protein fis. Phys. Rev. Lett. 2005, 95, 208101. [Google Scholar] [CrossRef] [PubMed]
- Dame, R.T.; Luijsterburg, M.S.; Krin, E.; Bertin, P.N.; Wagner, R.; Wuite, G.J. DNA bridging: A property shared among H-NS-like proteins. J. Bacteriol. 2005, 187, 1845–1848. [Google Scholar] [CrossRef] [PubMed]
- Luijsterburg, M.S.; White, M.F.; van Driel, R.; Dame, R.T. The major architects of chromatin: Architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem Mol. Biol. 2008, 43, 393–418. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, G.W.; Chen, C.; Xie, X.S.; Zhuang, X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science 2011, 333, 1445–1449. [Google Scholar] [CrossRef] [PubMed]
- Claret, L.; Rouviere-Yaniv, J. Variation in hu composition during growth of escherichia coli: The heterodimer is required for long term survival. J. Mol. Biol. 1997, 273, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Azam, T.A.; Ishihama, A. Twelve species of the nucleoid-associated protein from escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999, 274, 33105–33113. [Google Scholar] [CrossRef] [PubMed]
- Grainger, D.C. Structure and function of bacterial H-NS protein. Biochem. Soc. Trans. 2016, 44, 1561–1569. [Google Scholar] [CrossRef] [PubMed]
- Dame, R.T.; Wyman, C.; Goosen, N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic. Acids Res. 2000, 28, 3504–3510. [Google Scholar] [CrossRef] [PubMed]
- Amit, R.; Oppenheim, A.B.; Stavans, J. Single molecule elasticity measurements: A biophysical approach to bacterial nucleoid organization. Biophys. J. 2004, 87, 1392–1393. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; van der Valk, R.A.; Dame, R.T.; Roos, W.H.; Wuite, G.J.L. Probing the mechanical stability of bridged DNA-H-NS protein complexes by single-molecule afm pulling. Sci. Rep. 2017, 7, 15275. [Google Scholar] [CrossRef] [PubMed]
- Esposito, D.; Petrovic, A.; Harris, R.; Ono, S.; Eccleston, J.F.; Mbabaali, A.; Haq, I.; Higgins, C.F.; Hinton, J.C.; Driscoll, P.C.; et al. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J. Mol. Biol. 2002, 324, 841–850. [Google Scholar] [CrossRef]
- Lucchini, S.; Rowley, G.; Goldberg, M.D.; Hurd, D.; Harrison, M.; Hinton, J.C. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006, 2, e81. [Google Scholar] [CrossRef]
- Singh, S.S.; Singh, N.; Bonocora, R.P.; Fitzgerald, D.M.; Wade, J.T.; Grainger, D.C. Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev. 2014, 28, 214–219. [Google Scholar] [CrossRef] [Green Version]
- Rangarajan, A.A.; Schnetz, K. Interference of transcription across H-NS binding sites and repression by H-NS. Mol. Microbiol. 2018, 108, 226–239. [Google Scholar] [CrossRef] [Green Version]
- Wade, J.T.; Grainger, D.C. Waking the neighbours: Disruption of H-NS repression by overlapping transcription. Mol. Microbiol. 2018, 108, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Dame, R.T.; Tark-Dame, M. Bacterial chromatin: Converging views at different scales. Curr. Opin. Cell Biol. 2016, 40, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Le, T.B.; Imakaev, M.V.; Mirny, L.A.; Laub, M.T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 2013, 342, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Marbouty, M.; Koszul, R. Metagenome analysis exploiting high-throughput chromosome conformation capture (3c) data. Trends Genet. 2015, 31, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Atlung, T.; Ingmer, H. H-NS: A modulator of environmentally regulated gene expression. Mol. Microbiol. 1997, 24, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Talukder, A.; Ishihama, A. Growth phase dependent changes in the structure and protein composition of nucleoid in escherichia coli. Sci. China Life Sci. 2015, 58, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Busslinger, G.A.; Stocsits, R.R.; van der Lelij, P.; Axelsson, E.; Tedeschi, A.; Galjart, N.; Peters, J.M. Cohesin is positioned in mammalian genomes by transcription, ctcf and wapl. Nature 2017, 544, 503–507. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Satoh, M.; Kuroiwa, T. Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp. Cell Res. 1991, 196, 137–140. [Google Scholar] [CrossRef]
- Legros, F.; Malka, F.; Frachon, P.; Lombes, A.; Rojo, M. Organization and dynamics of human mitochondrial DNA. J. Cell Sci. 2004, 117, 2653–2662. [Google Scholar] [CrossRef] [Green Version]
- Mignotte, B.; Barat, M. Characterization of a xenopus laevis mitochondrial protein with a high affinity for supercoiled DNA. Nucleic Acids Res. 1986, 14, 5969–5980. [Google Scholar] [CrossRef] [PubMed]
- Alam, T.I.; Kanki, T.; Muta, T.; Ukaji, K.; Abe, Y.; Nakayama, H.; Takio, K.; Hamasaki, N.; Kang, D. Human mitochondrial DNA is packaged with tfam. Nucleic Acids Res. 2003, 31, 1640–1645. [Google Scholar] [CrossRef] [PubMed]
- Bogenhagen, D.F.; Rousseau, D.; Burke, S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008, 283, 3665–3675. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, B.A.; Newman, S.M.; Hallberg, R.L.; Slaughter, C.A.; Perlman, P.S.; Butow, R.A. In organello formaldehyde crosslinking of proteins to mtdna: Identification of bifunctional proteins. Proc. Natl. Acad Sci. USA 2000, 97, 7772–7777. [Google Scholar] [CrossRef] [PubMed]
- Rajala, N.; Hensen, F.; Wessels, H.J.; Ives, D.; Gloerich, J.; Spelbrink, J.N. Whole cell formaldehyde cross-linking simplifies purification of mitochondrial nucleoids and associated proteins involved in mitochondrial gene expression. PLoS ONE 2015, 10, e0116726. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Cooper, H.M.; Reyes, A.; Di Re, M.; Kazak, L.; Wood, S.R.; Mao, C.C.; Fearnley, I.M.; Walker, J.E.; Holt, I.J. Human c4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res. 2012, 40, 6097–6108. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bogenhagen, D.F. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006, 281, 25791–25802. [Google Scholar] [CrossRef] [PubMed]
- Hensen, F.; Cansiz, S.; Gerhold, J.M.; Spelbrink, J.N. To be or not to be a nucleoid protein: A comparison of mass-spectrometry based approaches in the identification of potential mtdna-nucleoid associated proteins. Biochimie 2014, 100, 219–226. [Google Scholar] [CrossRef]
- Han, S.; Udeshi, N.D.; Deerinck, T.J.; Svinkina, T.; Ellisman, M.H.; Carr, S.A.; Ting, A.Y. Proximity biotinylation as a method for mapping proteins associated with mtdna in living cells. Cell Chem. Biol. 2017, 24, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Dlaskova, A.; Engstova, H.; Spacek, T.; Kahancova, A.; Pavluch, V.; Smolkova, K.; Spackova, J.; Bartos, M.; Hlavata, L.P.; Jezek, P. 3d super-resolution microscopy reflects mitochondrial cristae alternations and mtdna nucleoid size and distribution. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Kukat, C.; Wurm, C.A.; Spahr, H.; Falkenberg, M.; Larsson, N.G.; Jakobs, S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtdna. Proc. Natl. Acad. Sci. USA 2011, 108, 13534–13539. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.A.; Tkachuk, A.N.; Shtengel, G.; Kopek, B.G.; Bogenhagen, D.F.; Hess, H.F.; Clayton, D.A. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol. Cell. Biol. 2011, 31, 4994–5010. [Google Scholar] [CrossRef] [PubMed]
- Kukat, C.; Davies, K.M.; Wurm, C.A.; Spahr, H.; Bonekamp, N.A.; Kuhl, I.; Joos, F.; Polosa, P.L.; Park, C.B.; Posse, V.; et al. Cross-strand binding of tfam to a single mtdna molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. USA 2015, 112, 11288–11293. [Google Scholar] [CrossRef] [PubMed]
- Alan, L.; Spacek, T.; Jezek, P. Delaunay algorithm and principal component analysis for 3d visualization of mitochondrial DNA nucleoids by biplane fpalm/dstorm. Eur. Biophys. J. 2016, 45, 443–461. [Google Scholar] [CrossRef] [PubMed]
- Spacek, T.; Pavluch, V.; Alan, L.; Capkova, N.; Engstova, H.; Dlaskova, A.; Berkova, Z.; Saudek, F.; Jezek, P. Nkx6.1 decline accompanies mitochondrial DNA reduction but subtle nucleoid size decrease in pancreatic islet beta-cells of diabetic goto kakizaki rats. Sci. Rep. 2017, 7, 15674. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, I. Organization and dynamics of yeast mitochondrial nucleoids. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 339–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendich, A.J. The end of the circle for yeast mitochondrial DNA. Mol. Cell 2010, 39, 831–832. [Google Scholar] [CrossRef]
- Diffley, J.F.; Stillman, B. A close relative of the nuclear, chromosomal high-mobility group protein hmg1 in yeast mitochondria. Proc. Natl. Acad. Sci. USA 1991, 88, 7864–7868. [Google Scholar] [CrossRef] [PubMed]
- Newman, S.M.; Zelenaya-Troitskaya, O.; Perlman, P.S.; Butow, R.A. Analysis of mitochondrial DNA nucleoids in wild-type and a mutant strain of saccharomyces cerevisiae that lacks the mitochondrial hmg box protein abf2p. Nucleic Acids Res. 1996, 24, 386–393. [Google Scholar] [CrossRef]
- Brewer, L.R.; Friddle, R.; Noy, A.; Baldwin, E.; Martin, S.S.; Corzett, M.; Balhorn, R.; Baskin, R.J. Packaging of single DNA molecules by the yeast mitochondrial protein abf2p. Biophys. J. 2003, 85, 2519–2524. [Google Scholar] [CrossRef]
- Friddle, R.W.; Klare, J.E.; Martin, S.S.; Corzett, M.; Balhorn, R.; Baldwin, E.P.; Baskin, R.J.; Noy, A. Mechanism of DNA compaction by yeast mitochondrial protein abf2p. Biophys. J. 2004, 86, 1632–1639. [Google Scholar] [CrossRef]
- Chakraborty, A.; Lyonnais, S.; Battistini, F.; Hospital, A.; Medici, G.; Prohens, R.; Orozco, M.; Vilardell, J.; Sola, M. DNA structure directs positioning of the mitochondrial genome packaging protein abf2p. Nucleic Acids Res. 2017, 45, 951–967. [Google Scholar] [CrossRef] [PubMed]
- Puhl, H.L.; Behe, M.J. Poly(da).Poly(dt) forms very stable nucleosomes at higher temperatures. J. Mol. Biol. 1995, 245, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, C.M.; Falkenberg, M.; Larsson, N.G. Maintenance and expression of mammalian mitochondrial DNA. Annu Rev. Biochem. 2016, 85, 133–160. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Dierckx, A.; Wanrooij, P.H.; Wanrooij, S.; Larsson, N.G.; Wilhelmsson, L.M.; Falkenberg, M.; Gustafsson, C.M. Mammalian transcription factor a is a core component of the mitochondrial transcription machinery. Proc. Natl. Acad. Sci. USA 2012, 109, 16510–16515. [Google Scholar] [CrossRef]
- Ngo, H.B.; Kaiser, J.T.; Chan, D.C. The mitochondrial transcription and packaging factor tfam imposes a u-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 2011, 18, 1290–1296. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Cosials, A.; Sidow, J.F.; Jimenez-Menendez, N.; Fernandez-Millan, P.; Montoya, J.; Jacobs, H.T.; Coll, M.; Bernado, P.; Sola, M. Human mitochondrial transcription factor a induces a u-turn structure in the light strand promoter. Nat. Struct Mol. Biol. 2011, 18, 1281–1289. [Google Scholar] [CrossRef]
- Falkenberg, M.; Gaspari, M.; Rantanen, A.; Trifunovic, A.; Larsson, N.G.; Gustafsson, C.M. Mitochondrial transcription factors b1 and b2 activate transcription of human mtdna. Nat. Genet. 2002, 31, 289–294. [Google Scholar] [CrossRef]
- Hillen, H.S.; Morozov, Y.I.; Sarfallah, A.; Temiakov, D.; Cramer, P. Structural basis of mitochondrial transcription initiation. Cell 2017, 171, 1072–1081. [Google Scholar] [CrossRef]
- Chimienti, G.; Picca, A.; Sirago, G.; Fracasso, F.; Calvani, R.; Bernabei, R.; Russo, F.; Carter, C.S.; Leeuwenburgh, C.; Pesce, V.; et al. Increased tfam binding to mtdna damage hot spots is associated with mtdna loss in aged rat heart. Free Radic Biol. Med. 2018, 124, 447–453. [Google Scholar] [CrossRef]
- Brown, T.A.; Tkachuk, A.N.; Clayton, D.A. Mitochondrial transcription factor a (tfam) binds to rna containing 4-way junctions and mitochondrial trna. PLoS ONE 2015, 10, e0142436. [Google Scholar] [CrossRef] [PubMed]
- Lyonnais, S.; Tarres-Sole, A.; Rubio-Cosials, A.; Cuppari, A.; Brito, R.; Jaumot, J.; Gargallo, R.; Vilaseca, M.; Silva, C.; Granzhan, A.; et al. The human mitochondrial transcription factor a is a versatile g-quadruplex binding protein. Sci. Rep. 2017, 7, 43992. [Google Scholar] [CrossRef] [PubMed]
- Ngo, H.B.; Lovely, G.A.; Phillips, R.; Chan, D.C. Distinct structural features of tfam drive mitochondrial DNA packaging versus transcriptional activation. Nat. Commun 2014, 5, 3077. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Cosials, A.; Battistini, F.; Gansen, A.; Cuppari, A.; Bernado, P.; Orozco, M.; Langowski, J.; Toth, K.; Sola, M. Protein flexibility and synergy of hmg domains underlie u-turn bending of DNA by tfam in solution. Biophys. J. 2018, 114, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Malarkey, C.S.; Bestwick, M.; Kuhlwilm, J.E.; Shadel, G.S.; Churchill, M.E. Transcriptional activation by mitochondrial transcription factor a involves preferential distortion of promoter DNA. Nucleic Acids Res. 2012, 40, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Farge, G.; Laurens, N.; Broekmans, O.D.; van den Wildenberg, S.M.; Dekker, L.C.; Gaspari, M.; Gustafsson, C.M.; Peterman, E.J.; Falkenberg, M.; Wuite, G.J. Protein sliding and DNA denaturation are essential for DNA organization by human mitochondrial transcription factor a. Nat. Commun. 2012, 3, 1013. [Google Scholar] [CrossRef]
- Traverso, J.J.; Manoranjan, V.S.; Bishop, A.R.; Rasmussen, K.O.; Voulgarakis, N.K. Allostery through protein-induced DNA bubbles. Sci. Rep. 2015, 5, 9037. [Google Scholar] [CrossRef]
- Kaufman, B.A.; Durisic, N.; Mativetsky, J.M.; Costantino, S.; Hancock, M.A.; Grutter, P.; Shoubridge, E.A. The mitochondrial transcription factor tfam coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell 2007, 18, 3225–3236. [Google Scholar] [CrossRef]
- Gangelhoff, T.A.; Mungalachetty, P.S.; Nix, J.C.; Churchill, M.E. Structural analysis and DNA binding of the hmg domains of the human mitochondrial transcription factor a. Nucleic Acids Res. 2009, 37, 3153–3164. [Google Scholar] [CrossRef]
- Wong, T.S.; Rajagopalan, S.; Freund, S.M.; Rutherford, T.J.; Andreeva, A.; Townsley, F.M.; Petrovich, M.; Fersht, A.R. Biophysical characterizations of human mitochondrial transcription factor a and its binding to tumor suppressor p53. Nucleic Acids Res. 2009, 37, 6765–6783. [Google Scholar] [CrossRef]
- Kasashima, K.; Endo, H. Interaction of human mitochondrial transcription factor a in mitochondria: Its involvement in the dynamics of mitochondrial DNA nucleoids. Genes Cells 2015, 20, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Ohgaki, K.; Kanki, T.; Fukuoh, A.; Kurisaki, H.; Aoki, Y.; Ikeuchi, M.; Kim, S.H.; Hamasaki, N.; Kang, D. The c-terminal tail of mitochondrial transcription factor a markedly strengthens its general binding to DNA. J. Biochem. 2007, 141, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Falkenberg, M. Mitochondrial DNA replication in mammalian cells: Overview of the pathway. Essays Biochem. 2018, 62, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Krasich, R.; Copeland, W.C. DNA polymerases in the mitochondria: A critical review of the evidence. Front. Biosci. 2017, 22, 692–709. [Google Scholar]
- Ziebarth, T.D.; Gonzalez-Soltero, R.; Makowska-Grzyska, M.M.; Nunez-Ramirez, R.; Carazo, J.M.; Kaguni, L.S. Dynamic effects of cofactors and DNA on the oligomeric state of human mitochondrial DNA helicase. J. Biol. Chem. 2010, 285, 14639–14647. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Millan, P.; Lazaro, M.; Cansiz-Arda, S.; Gerhold, J.M.; Rajala, N.; Schmitz, C.A.; Silva-Espina, C.; Gil, D.; Bernado, P.; Valle, M.; et al. The hexameric structure of the human mitochondrial replicative helicase twinkle. Nucleic Acids Res. 2015, 43, 4284–4295. [Google Scholar] [CrossRef] [PubMed]
- Peter, B.; Farge, G.; Pardo-Hernandez, C.; Tangefjord, S.; Falkenberg, M. Structural basis for adpeo-causing mutations in the mitochondrial twinkle helicase. Hum. Mol. Genet. 2019, 28, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Miralles Fuste, J.; Shi, Y.; Wanrooij, S.; Zhu, X.; Jemt, E.; Persson, O.; Sabouri, N.; Gustafsson, C.M.; Falkenberg, M. In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet. 2014, 10, e1004832. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, T.J.; Nadalutti, C.A.; Motori, E.; Sommerville, E.W.; Gorman, G.S.; Basu, S.; Hoberg, E.; Turnbull, D.M.; Chinnery, P.F.; Larsson, N.G.; et al. Topoisomerase 3alpha is required for decatenation and segregation of human mtdna. Mol. Cell 2018, 69, 9–23. [Google Scholar] [CrossRef]
- Posse, V.; Al-Behadili, A.; Uhler, J.P.; Clausen, A.R.; Reyes, A.; Zeviani, M.; Falkenberg, M.; Gustafsson, C.M. Rnase h1 directs origin-specific initiation of DNA replication in human mitochondria. PLoS Genet. 2019, 15, e1007781. [Google Scholar] [CrossRef]
- Agaronyan, K.; Morozov, Y.I.; Anikin, M.; Temiakov, D. Mitochondrial biology. Replication-transcription switch in human mitochondria. Science 2015, 347, 548–551. [Google Scholar] [CrossRef] [PubMed]
- Posse, V.; Shahzad, S.; Falkenberg, M.; Hallberg, B.M.; Gustafsson, C.M. Tefm is a potent stimulator of mitochondrial transcription elongation in vitro. Nucleic Acids Res. 2015, 43, 2615–2624. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Koolmeister, C.; Misic, J.; Siira, S.; Kuhl, I.; Silva Ramos, E.; Miranda, M.; Jiang, M.; Posse, V.; Lytovchenko, O.; et al. Tefm regulates both transcription elongation and rna processing in mitochondria. EMBO Rep. 2019. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, E.; Sclavi, B. Gene regulation by H-NS as a function of growth conditions depends on chromosomal position in Escherichia coli. G3 2015, 5, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, I.J.; Greene, E.C. Molecular traffic jams on DNA. Annu Rev. Biophys. 2013, 42, 241–263. [Google Scholar] [CrossRef] [PubMed]
- Kucej, M.; Kucejova, B.; Subramanian, R.; Chen, X.J.; Butow, R.A. Mitochondrial nucleoids undergo remodeling in response to metabolic cues. J. Cell Sci. 2008, 121, 1861–1868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelenaya-Troitskaya, O.; Newman, S.M.; Okamoto, K.; Perlman, P.S.; Butow, R.A. Functions of the high mobility group protein, abf2p, in mitochondrial DNA segregation, recombination and copy number in saccharomyces cerevisiae. Genetics 1998, 148, 1763–1776. [Google Scholar]
- Farge, G.; Mehmedovic, M.; Baclayon, M.; van den Wildenberg, S.M.; Roos, W.H.; Gustafsson, C.M.; Wuite, G.J.; Falkenberg, M. In vitro-reconstituted nucleoids can block mitochondrial DNA replication and transcription. Cell Rep. 2014, 8, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Wai, T.; Teoli, D.; Shoubridge, E.A. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat. Genet. 2008, 40, 1484–1488. [Google Scholar] [CrossRef]
- Dinardo, M.M.; Musicco, C.; Fracasso, F.; Milella, F.; Gadaleta, M.N.; Gadaleta, G.; Cantatore, P. Acetylation and level of mitochondrial transcription factor a in several organs of young and old rats. Biochem. Biophys. Res. Commun. 2003, 301, 187–191. [Google Scholar] [CrossRef]
- Lu, B.; Lee, J.; Nie, X.; Li, M.; Morozov, Y.I.; Venkatesh, S.; Bogenhagen, D.F.; Temiakov, D.; Suzuki, C.K. Phosphorylation of human tfam in mitochondria impairs DNA binding and promotes degradation by the aaa+ lon protease. Mol. Cell 2013, 49, 121–132. [Google Scholar] [CrossRef] [PubMed]
- King, G.A.; Hashemi Shabestari, M.; Taris, K.H.; Pandey, A.K.; Venkatesh, S.; Thilagavathi, J.; Singh, K.; Krishna Koppisetti, R.; Temiakov, D.; Roos, W.H.; et al. Acetylation and phosphorylation of human tfam regulate tfam-DNA interactions via contrasting mechanisms. Nucleic Acids Res. 2018, 46, 3633–3642. [Google Scholar] [CrossRef] [PubMed]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farge, G.; Falkenberg, M. Organization of DNA in Mammalian Mitochondria. Int. J. Mol. Sci. 2019, 20, 2770. https://doi.org/10.3390/ijms20112770
Farge G, Falkenberg M. Organization of DNA in Mammalian Mitochondria. International Journal of Molecular Sciences. 2019; 20(11):2770. https://doi.org/10.3390/ijms20112770
Chicago/Turabian StyleFarge, Géraldine, and Maria Falkenberg. 2019. "Organization of DNA in Mammalian Mitochondria" International Journal of Molecular Sciences 20, no. 11: 2770. https://doi.org/10.3390/ijms20112770
APA StyleFarge, G., & Falkenberg, M. (2019). Organization of DNA in Mammalian Mitochondria. International Journal of Molecular Sciences, 20(11), 2770. https://doi.org/10.3390/ijms20112770