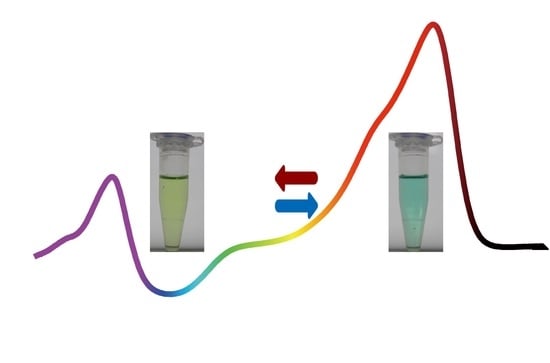
Protein Engineering of Dual-Cys Cyanobacteriochrome AM1_1186g2 for Biliverdin Incorporation and Far-Red/Blue Reversible Photoconversion
Abstract
:1. Introduction
2. Results
2.1. RCAP: Efficient BV Incorporation
2.2. KCAP: Improvement in Protein Stability
2.3. KCAP_QV: Improvement in Holoprotein Expression
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Expression and Purification of His-Tagged AM1_1186g2 Variants
4.3. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Zinc-Dependent Fluorescence Gel Assay
4.4. Western Blotting Assay
4.5. Spectrometry and Measurement of Photoconversion Kinetics
4.6. HPLC Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kehoe, D.M.; Grossman, A.R. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 1996, 273, 1409–1412. [Google Scholar] [CrossRef] [PubMed]
- Hirose, Y.; Narikawa, R.; Katayama, M.; Ikeuchi, M. Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proc. Natl. Acad. Sci. USA 2010, 107, 8854–8859. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, S.; Suzuki, F.; Fujita, H.; Geng, X.X.; Ikeuchi, M. Novel putative photoreceptor and regulatory genes required for the positive phototactic movement of the unicellular motile cyanobacterium synechocystis sp. PCC 6803. Plant. Cell Physiol. 2000, 41, 1299–1304. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, G.; Nomura, R.; Shimada, T.; Ni-Ni-Win; Narikawa, R.; Ikeuchi, M. Cyanobacteriochrome SesA is a diguanylate cyclase that induces cell aggregation in Thermosynechococcus. J. Biol. Chem. 2014, 289, 24801–24809. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, G.; Ni-Ni-Win; Narikawa, R.; Ikeuchi, M. Three cyanobacteriochromes work together to form a light color-sensitive input system for c-di-GMP signaling of cell aggregation. Proc. Natl. Acad. Sci. USA 2015, 112, 8082–8087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, F.; Zhang, S.; Rockwell, N.C.; Martin, S.S.; Lagarias, J.C.; Bryant, D.A. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 2014, 345, 1312–1317. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Gan, F.; Shen, G.; Bryant, D.A. RfpA, RfpB, and RfpC are the master control elements of far-red light photoacclimation (FaRLiP). Front. Microbiol. 2015, 6, 1303. [Google Scholar] [CrossRef]
- Narikawa, R.; Suzuki, F.; Yoshihara, S.; Higashi, S.-I.; Watanabe, M.; Ikeuchi, M. Novel photosensory two-component system (PixA-NixB-NixC) involved in the regulation of positive and negative phototaxis of cyanobacterium synechocystis sp. PCC 6803. Plant. Cell Physiol. 2011, 52, 2214–2224. [Google Scholar] [CrossRef]
- Song, J.-Y.; Cho, H.S.; Cho, J.-I.; Jeon, J.-S.; Lagarias, J.C.; Park, Y.-I. Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 2011, 108, 10780–10785. [Google Scholar] [CrossRef]
- Wiltbank, L.B.; Kehoe, D.M. Two cyanobacterial photoreceptors regulate photosynthetic light harvesting by sensing teal, green, yellow, and red light. MBio 2016, 7, e02130-15. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Lagarias, J.C. A brief history of phytochromes. Chemphyschem 2010, 11, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Anders, K.; Essen, L.-O. The family of phytochrome-like photoreceptors: Diverse, complex and multi-colored, but very useful. Curr. Opin. Struct. Biol. 2015, 35, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Fushimi, K.; Narikawa, R. Cyanobacteriochromes: Photoreceptors covering the entire UV-to-visible spectrum. Curr. Opin. Struct. Biol. 2019, 57, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Ishizuka, T. Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 2008, 7, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, S.; Katayama, M.; Geng, X.; Ikeuchi, M. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant. Cell Physiol. 2004, 45, 1729–1737. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Njuguna, S.L.; Roberts, L.; Castillo, E.; Parson, V.L.; Dwojak, S.; Lagarias, J.C.; Spiller, S.C. A second conserved GAF domain cysteine is required for the blue/green photoreversibility of cyanobacteriochrome Tlr0924 from thermosynechococcus elongatus. Biochemistry 2008, 47, 7304–7316. [Google Scholar] [CrossRef]
- Narikawa, R.; Fukushima, Y.; Ishizuka, T.; Itoh, S.; Ikeuchi, M. A novel photoactive GAF domain of cyanobacteriochrome AnPixJ that shows reversible green/red photoconversion. J. Mol. Biol. 2008, 380, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, R.; Kohchi, T.; Ikeuchi, M. Characterization of the photoactive GAF domain of the CikA homolog (SyCikA, Slr1969) of the cyanobacterium synechocystis sp. PCC 6803. Photochem. Photobiol. Sci. 2008, 7, 1253–1259. [Google Scholar] [CrossRef]
- Hirose, Y.; Shimada, T.; Narikawa, R.; Katayama, M.; Ikeuchi, M. Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc. Natl. Acad. Sci. USA 2008, 105, 9528–9533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rockwell, N.C.; Martin, S.S.; Gulevich, A.G.; Lagarias, J.C. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry 2012, 51, 1449–1463. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Hua, H.-H.; Chen, Y.; Liu, B.-B.; Krämer, A.L.; Scheer, H.; Zhao, K.-H.; Zhou, M. A rising tide of blue-absorbing biliprotein photoreceptors: Characterization of seven such bilin-binding GAF domains in Nostoc sp. PCC7120. FEBS J. 2012, 279, 4095–4108. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, G.; Hirose, Y.; Narikawa, R.; Ikeuchi, M. Thiol-based photocycle of the blue and teal light-sensing cyanobacteriochrome Tlr1999. Biochemistry 2012, 51, 3050–3058. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Martin, S.S.; Feoktistova, K.; Lagarias, J.C. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc. Natl. Acad. Sci. USA 2011, 108, 11854–11859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narikawa, R.; Enomoto, G.; Ni-Ni-Win; Fushimi, K.; Ikeuchi, M. A new type of dual-Cys cyanobacteriochrome GAF domain found in cyanobacterium acaryochloris marina, which has an unusual red/blue reversible photoconversion cycle. Biochemistry 2014, 53, 5051–5059. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Lagarias, J.C. Identification of cyanobacteriochromes detecting far-red light. Biochemistry 2016, 55, 3907–3919. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Lagarias, J.C. There and back again: Loss and reacquisition of two-Cys photocycles in cyanobacteriochromes. Photochem. Photobiol. 2017, 93, 741–754. [Google Scholar] [CrossRef]
- Fushimi, K.; Rockwell, N.C.; Enomoto, G.; Ni-Ni-Win; Martin, S.S.; Gan, F.; Bryant, D.A.; Ikeuchi, M.; Lagarias, J.C.; Narikawa, R. Cyanobacteriochrome photoreceptors lacking the canonical Cys residue. Biochemistry 2016, 55, 6981–6995. [Google Scholar] [CrossRef]
- Narikawa, R.; Ishizuka, T.; Muraki, N.; Shiba, T.; Kurisu, G.; Ikeuchi, M. Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, 918–923. [Google Scholar] [CrossRef]
- Burgie, E.S.; Walker, J.M.; Phillips, G.N., Jr.; Vierstra, R.D. A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCF-cyanobacteriochromes. Structure 2013, 21, 88–97. [Google Scholar] [CrossRef]
- Cornilescu, C.C.; Cornilescu, G.; Burgie, E.S.; Markley, J.L.; Ulijasz, A.T.; Vierstra, R.D. Dynamic structural changes underpin photoconversion of a blue/green cyanobacteriochrome between its dark and photoactivated states. J. Biol. Chem. 2014, 289, 3055–3065. [Google Scholar] [CrossRef]
- Ishizuka, T.; Kamiya, A.; Suzuki, H.; Narikawa, R.; Noguchi, T.; Kohchi, T.; Inomata, K.; Ikeuchi, M. The cyanobacteriochrome, TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry 2011, 50, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Lagarias, J.C. Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochemistry 2012, 51, 3576–3585. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Gulevich, A.G.; Lagarias, J.C. Conserved phenylalanine residues are required for blue-shifting of cyanobacteriochrome photoproducts. Biochemistry 2014, 53, 3118–3130. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Fushimi, K.; Miyake, K.; Nakajima, T.; Oikawa, Y.; Enomoto, G.; Sato, M.; Ikeuchi, M.; Narikawa, R. Molecular characterization of DXCF cyanobacteriochromes from the cyanobacterium acaryochloris marina identifies a blue-light power sensor. J. Biol. Chem. 2018, 293, 1713–1727. [Google Scholar] [CrossRef]
- Cho, S.M.; Jeoung, S.C.; Song, J.-Y.; Song, J.-J.; Park, Y.-I. Hydrophobic residues near the bilin chromophore-binding pocket modulate spectral tuning of insert-Cys subfamily cyanobacteriochromes. Sci. Rep. 2017, 7, 40576. [Google Scholar] [CrossRef] [PubMed]
- Narikawa, R.; Nakajima, T.; Aono, Y.; Fushimi, K.; Enomoto, G.; Ni-Ni-Win; Itoh, S.; Sato, M.; Ikeuchi, M. A biliverdin-binding cyanobacteriochrome from the chlorophyll d-bearing cyanobacterium acaryochloris marina. Sci. Rep. 2015, 5, 7950. [Google Scholar] [CrossRef]
- Fushimi, K.; Nakajima, T.; Aono, Y.; Yamamoto, T.; Ni-Ni-Win; Ikeuchi, M.; Sato, M.; Narikawa, R. Photoconversion and fluorescence properties of a red/green-type cyanobacteriochrome AM1_C0023g2 that binds not only phycocyanobilin but also biliverdin. Front. Microbiol. 2016, 7, 588. [Google Scholar] [CrossRef] [PubMed]
- Fushimi, K.; Miyazaki, T.; Kuwasaki, Y.; Nakajima, T.; Yamamoto, T.; Suzuki, K.; Ueda, Y.; Miyake, K.; Takeda, Y.; Choi, J.-H.; et al. Rational conversion of chromophore selectivity of cyanobacteriochromes to accept mammalian intrinsic biliverdin. Proc. Natl. Acad. Sci. USA 2019, 116, 8301–8309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krumholz, A.; Shcherbakova, D.M.; Xia, J.; Wang, L.V.; Verkhusha, V.V. Multicontrast photoacoustic in vivo imaging using near-infrared fluorescent proteins. Sci. Rep. 2014, 4, 3939. [Google Scholar] [CrossRef] [Green Version]
- Subach, F.V.; Zhang, L.; Gadella, T.W.; Gurskaya, N.G.; Lukyanov, K.A.; Verkhusha, V.V. Red fluorescent protein with reversibly photoswitchable absorbance for photochromic FRET. Chem. Biol. 2010, 17, 745–755. [Google Scholar] [CrossRef]




| Name | Dark State (λmax) | Photoproduct (λmax) | SAR |
|---|---|---|---|
| WT | 687 nm | 398 nm | 0.11 |
| PR | - | - | 0.03 |
| RCAP | 691 nm | 398 nm | 0.65 |
| CAP | 691 nm | 398 nm | 0.40 |
| KCAP | 691 nm | 398 nm | 0.76 |
| KCAP_V2503Q | 690 nm | 398 nm | 0.61 |
| KCAP_N2504E | 691 nm | 398 nm | 0.18 |
| KCAP_N2508G | 691 nm | 398 nm | 0.48 |
| KCAP_Q2517V | 691 nm | 398 nm | 1.09 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuwasaki, Y.; Miyake, K.; Fushimi, K.; Takeda, Y.; Ueda, Y.; Nakajima, T.; Ikeuchi, M.; Sato, M.; Narikawa, R. Protein Engineering of Dual-Cys Cyanobacteriochrome AM1_1186g2 for Biliverdin Incorporation and Far-Red/Blue Reversible Photoconversion. Int. J. Mol. Sci. 2019, 20, 2935. https://doi.org/10.3390/ijms20122935
Kuwasaki Y, Miyake K, Fushimi K, Takeda Y, Ueda Y, Nakajima T, Ikeuchi M, Sato M, Narikawa R. Protein Engineering of Dual-Cys Cyanobacteriochrome AM1_1186g2 for Biliverdin Incorporation and Far-Red/Blue Reversible Photoconversion. International Journal of Molecular Sciences. 2019; 20(12):2935. https://doi.org/10.3390/ijms20122935
Chicago/Turabian StyleKuwasaki, Yuto, Keita Miyake, Keiji Fushimi, Yuka Takeda, Yoshibumi Ueda, Takahiro Nakajima, Masahiko Ikeuchi, Moritoshi Sato, and Rei Narikawa. 2019. "Protein Engineering of Dual-Cys Cyanobacteriochrome AM1_1186g2 for Biliverdin Incorporation and Far-Red/Blue Reversible Photoconversion" International Journal of Molecular Sciences 20, no. 12: 2935. https://doi.org/10.3390/ijms20122935
APA StyleKuwasaki, Y., Miyake, K., Fushimi, K., Takeda, Y., Ueda, Y., Nakajima, T., Ikeuchi, M., Sato, M., & Narikawa, R. (2019). Protein Engineering of Dual-Cys Cyanobacteriochrome AM1_1186g2 for Biliverdin Incorporation and Far-Red/Blue Reversible Photoconversion. International Journal of Molecular Sciences, 20(12), 2935. https://doi.org/10.3390/ijms20122935