Low Overnight Temperature-Induced Gibberellin Accumulation Increases Locule Number in Tomato
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Analysis of Tomato Fruit at Different Night Temperatures
2.2. Overview of Messenger RNA (mRNA) Sequencing Data
2.3. Overall Identification and Functional Annotation of Differentially Expressed Genes (DEGs)
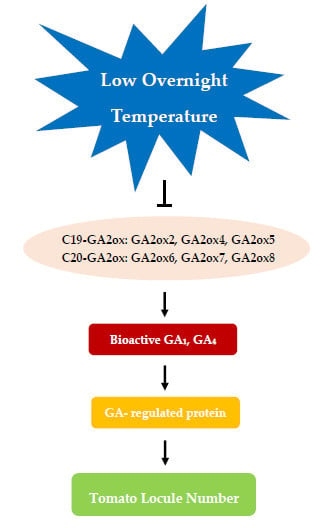
2.4. Expression Patterns and Endogenous Levels of Gibberellins
2.5. Validation of DEGs by qRT-PCR
2.6. Exogenous Gibberellin Applications Changed the Number of Tomato Locules
3. Discussion
3.1. Low Temperature Induced Multi-Locule Fruit Formation
3.2. Low Temperature Induced Accumulation of Bioactive GA1 and GA4
3.3. Effect of Gibberellins on the Formation of Tomato Locules
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Treatments
4.2. Phenotypic Analysis of Tomato Fruit
4.3. RNA Extraction and Transcriptome Sequencing
4.4. Data Analysis
4.5. qRT–PCR Analysis
4.6. Quantification of Phytohormones
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhu, J.; Dong, C.; Zhu, J. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 2007, 10, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, P.; Yan, Y.; Bao, C.; Li, X.; Wang, L.; Shen, X.; Li, H.; Liu, X.; Niu, C. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 2018, 218, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.; Clement, C.; Barka, E.A. Physiological and molecular changes in plants grown at low temperatures. Planta 2012, 235, 1091–1105. [Google Scholar] [CrossRef]
- Sairam, R.K.; Tyagi, A. Physiology and molecular biology of salinity stress tolerance in plants. Curr. Sci. 2004, 86, 407–421. [Google Scholar]
- Shinozaki, K.; Yamaguchishinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Lozano, R.; Angosto, T.; Gomez, P.; Payan, C.; Capel, J.; Huijser, P.; Salinas, J.; Martinezzapater, J.M. Tomato Flower Abnormalities Induced by Low Temperatures Are Associated with Changes of Expression of MADS-Box Genes. Plant Physiol. 1998, 117, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Aloni, B.; Pressman, E.; Karni, L. The Effect of Fruit Load, Defoliation and Night Temperature on the Morphology of Pepper Flowers and on Fruit Shape. Ann. Bot. 1999, 83, 529–534. [Google Scholar] [CrossRef] [Green Version]
- Ma, N.; Chen, W.; Fan, T.; Tian, Y.; Zhang, S.; Zeng, D.; Li, Y. Low temperature-induced DNA hypermethylation attenuates expression of RhAG, an AGAMOUS homolog, and increases petal number in rose (Rosa hybrida). BMC Plant Biol. 2015, 15, 237. [Google Scholar] [CrossRef]
- Hong, J.H.; Savina, M.; Du, J.; Devendran, A.; Ramakanth, K.K.; Tian, X.; Sim, W.S.; Mironova, V.V.; Xu, J. A Sacrifice-for-Survival Mechanism Protects Root Stem Cell Niche from Chilling Stress. Cell 2017, 170, 102–113. [Google Scholar] [CrossRef]
- Bowman, J.L.; Eshed, Y. Formation and maintenance of the shoot apical meristem. Trends Plant Sci. 2000, 5, 110–115. [Google Scholar] [CrossRef]
- Holt, A.L.; Van Haperen, J.M.; Groot, E.P.; Laux, T. Signaling in shoot and flower meristems of Arabidopsis thaliana. Curr. Opin. Plant Biol. 2014, 17, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Galli, M.; Gallavotti, A. Expanding the Regulatory Network for Meristem Size in Plants. Trends Genet. 2016, 32, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Cong, B.; Barrero, L.; Tanksley, S.D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008, 40, 800–804. [Google Scholar] [CrossRef] [PubMed]
- Lozano, R.; Gimenez, E.; Cara, B.; Capel, J.; Angosto, T. Genetic analysis of reproductive development in tomato. Int. J. Dev. Biol. 2009, 53, 1635–1648. [Google Scholar] [CrossRef] [PubMed]
- Munos, S.; Ranc, N.; Botton, E.; Berard, A.; Rolland, S.; Duffe, P.; Carretero, Y.; Paslier, M.L.; Delalande, C.; Bouzayen, M. Increase in Tomato Locule Number Is Controlled by Two Single-Nucleotide Polymorphisms Located Near WUSCHEL. Plant Physiol. 2011, 156, 2244–2254. [Google Scholar] [CrossRef] [PubMed]
- Tanksley, S.D. The Genetic, Developmental, and Molecular Bases of Fruit Size and Shape Variation in Tomato. Plant Cell 2004, 16, S181–S189. [Google Scholar] [CrossRef]
- Lippman, Z.; Tanksley, S.D. Dissecting the genetic pathway to extreme fruit size in tomato using a cross between the small-fruited wild species Lycopersicon pimpinellifolium and L. esculentum var. Giant Heirloom. Genetics 2001, 158, 413–422. [Google Scholar] [PubMed]
- Li, Y.; Li, T.; Wang, D. Studies on the Inheritance of Locule Formation in Tomatoes (Lycopersicon esculentum Mill.). J. Genet. Genom. 2007, 34, 1028–1036. [Google Scholar] [CrossRef]
- Rodriguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480. [Google Scholar] [CrossRef]
- Barrero, L.; Tanksley, S.D. Evaluating the genetic basis of multiple-locule fruit in a broad cross section of tomato cultivars. Theor. Appl. Genet. 2004, 109, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Illaberenguer, E.; Van Houten, J.; Huang, Z.; Der Knaap, E.V. Rapid and reliable identification of tomato fruit weight and locule number loci by QTL-seq. Theor. Appl. Genet. 2015, 128, 1329–1342. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qi, M.; Sun, M.; Liu, Y.; Liu, Y.; Xu, T.; Li, Y.; Li, T. Tomato transcription factor SlWUS plays an important role in tomato flower and locule development. Front. Plant Sci. 2017, 8, 457. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Liberatore, K.L.; Macalister, C.A.; Huang, Z.; Chu, Y.H.; Jiang, K.; Brooks, C.; Ogawaohnishi, M.; Xiong, G.; Pauly, M. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015, 47, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, T.L. Regulation effects of exogenous gibberellin acid (GA3) on the formation of tomato (Solanum Lycoperscium) ovary locule and fasciated transcription. Afr. J. Biotechnol. 2012, 11, 13732–13738. [Google Scholar]
- Sawhney, V.K. The role of temperature and its relationship with gibberellic acid in the development of floral organs of tomato (Lycopersicon esculentum). Can. J. Bot. 1983, 61, 1258–1265. [Google Scholar] [CrossRef]
- Pressman, E.; Moshkovitch, H.; Rosenfeld, K.; Shaked, R.; Gamliel, B.; Aloni, B. Influence of low night temperatures on sweet pepper flower quality and the effect of repeated pollinations, with viable pollen, on fruit setting. J. Hortic. Sci. Biotech. 1998, 73, 131–136. [Google Scholar] [CrossRef]
- Asahira, T.; Hosoki, T.; Shinya, K. Regulation of low temperature-induced malformation of tomato fruit by plant growth regulators. J. Jpn. Soc. Hortic. Sci. 1982, 50, 468–474. [Google Scholar] [CrossRef]
- Urbanova, T.; Tarkowska, D.; Novak, O.; Hedden, P.; Strnad, M. Analysis of gibberellins as free acids by ultra performance liquid chromatography–tandem mass spectrometry. Talanta 2013, 112, 85–94. [Google Scholar] [CrossRef]
- Hirano, K.; Aya, K.; Hobo, T.; Sakakibara, H.; Kojima, M.; Shim, R.A.; Hasegawa, Y.; Ueguchitanaka, M.; Matsuoka, M. Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant Cell Physiol. 2008, 49, 1429–1450. [Google Scholar] [CrossRef]
- Chen, S.; Wang, X.; Zhang, L.; Lin, S.; Liu, D.; Wang, Q.; Cai, S.; El-Tanbouly, R.; Gan, L.; Wu, H.; et al. Identification and characterization of tomato gibberellin 2-oxidases (GA2oxs) and effects of fruit-specific SlGA2ox1 overexpression on fruit and seed growth and development. Hortic. Res. 2016, 3, 16059. [Google Scholar] [CrossRef] [PubMed]
- Martinezbello, L.; Moritz, T.; Lopezdiaz, I. Silencing C19-GA 2-oxidases induces parthenocarpic development and inhibits lateral branching in tomato plants. J. Exp. Bot. 2015, 66, 5897–5910. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Watanabe, M.; Shiogama, H.; Inoue, J.; Kimoto, M. Robust Arctic sea-ice influence on the frequent Eurasian cold winters in past decades. Nat. Geosci. 2014, 7, 869–873. [Google Scholar] [CrossRef]
- Overland, J.E.; Dethloff, K.; Francis, J.A.; Hall, R.J.; Hanna, E.; Kim, S.; Screen, J.A.; Shepherd, T.G.; Vihma, T. Nonlinear response of mid-latitude weather to the changing Arctic. Nat. Clim. Chang. 2016, 6, 992–999. [Google Scholar] [CrossRef] [Green Version]
- Shepherd, T.G. Effects of a warming Arctic. Science 2016, 353, 989–990. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M. Patterns in plant development (2nd edn). Trends Genet. 1990, 6, 97. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, R.; Zi, H.; Li, Y.; Cao, X.; Li, D.; Guo, L.; Tong, J.; Pan, Y.; Jiao, Y. AUXIN RESPONSE FACTOR3 Regulates Floral Meristem Determinacy by Repressing Cytokinin Biosynthesis and Signaling. Plant Cell 2018, 30, 324–346. [Google Scholar] [CrossRef]
- Beppu, K.; Ikeda, T.; Kataoka, I. Effect of high temperature exposure time during flower bud formation on the occurrence of double pistils in ‘Satohnishiki’ sweet cherry. Sci. Hortic. 2001, 87, 77–84. [Google Scholar] [CrossRef]
- Pan, I.L.; Mcquinn, R.; Giovannoni, J.J.; Irish, V.F. Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J. Exp. Bot. 2010, 61, 1795–1806. [Google Scholar] [CrossRef]
- Itkin, M.; Seybold, H.; Breitel, D.; Rogachev, I.; Meir, S.; Aharoni, A. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 2009, 60, 1081–1095. [Google Scholar] [CrossRef]
- Shang, M.; Wang, X.; Zhang, J.; Qi, X.; Ping, A.; Hou, L.; Xing, G.; Li, G.; Li, M. Genetic Regulation of GA Metabolism during Vernalization, Floral Bud Initiation and Development in Pak Choi (Brassica rapa ssp. chinensis Makino). Front. Plant Sci. 2017, 8, 1533. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ito, T.; Zhao, Y.; Peng, J.; Kumar, P.P.; Meyerowitz, E.M. Floral homeotic genes are targets of gibberellin signaling in flower development. Proc. Natl. Acad. Sci. USA 2004, 101, 7827–7832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shani, E.; Yanai, O.; Ori, N. The role of hormones in shoot apical meristem function. Curr. Opin. Plant Biol. 2006, 9, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Kobayashi, M.; Itoh, H.; Tagiri, A.; Kayano, T.; Tanaka, H.; Iwahori, S.; Matsuoka, M. Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 2001, 125, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, S.; Piazza, P.; Craft, J.; Hay, A.; Woolley, L.; Rieu, I.; Phillips, A.L.; Hedden, P.; Tsiantis, M. KNOX Action in Arabidopsis Is Mediated by Coordinate Regulation of Cytokinin and Gibberellin Activities. Curr. Biol. 2005, 15, 1560–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Torres-Garcia, J.; Latrasse, D.; Benhamed, M.; Schilderink, S.; Zhou, W.; Kulikova, O.; Hirt, H.; Bisseling, T. Plant-Specific Histone Deacetylases HDT1/2 Regulate GIBBERELLIN 2-OXIDASE2 Expression to Control Arabidopsis Root Meristem Cell Number. Plant Cell 2017, 29, 2183–2196. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential Expression of RNA-Seq Data at the Gene Level–The DESeq Package; European Molecular Biology Laboratory (EMBL): Heidelberg, Germany, 2012. [Google Scholar]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef]






| Samples | Total Raw Reads | Total Reads | Total Mapped (%) | Uniquely Mapped (%) |
|---|---|---|---|---|
| CK | 73,023,120 | 72,347,574 | 67,829,267 (93.75) | 65,060,425 (89.93) |
| T10-d10 | 64,078,977 | 63,600,058 | 60,112,344 (94.52) | 57,752,715 (90.81) |
| T15-d10 | 53,299,090 | 52,897,547 | 49,928,204 (94.39) | 47,913,998 (90.58) |
| T20-d10 | 52,736,947 | 52,392,032 | 49,551,460 (94.58) | 47,523,527 (90.71) |
| T10-d20 | 59,988,215 | 59,600,279 | 56,433,514 (94.69) | 54,114,303 (90.80) |
| T15-d20 | 53,972,546 | 53,397,506 | 49,836,432 (93.33) | 47,801,791 (89.52) |
| T20-d20 | 67,265,972 | 66,417,818 | 62,130,622 (93.55) | 59,583,787 (89.71) |
| Pathway ID | Pathway | Number of DEGs with Pathway Annotation | |||||
|---|---|---|---|---|---|---|---|
| T10-d10 vs. CK | T15-d10 vs. CK | T20-d10 vs. CK | T10-d20 vs. CK | T15-d20 vs. CK | T20-d20 vs. CK | ||
| ko04075 | Plant hormone signal transduction | 0 | 0 | 38 | 58 | 44 | 48 |
| ko00500 | Starch and sucrose metabolism | 4 | 6 | 0 | 37 | 33 | 31 |
| ko04626 | Plant-pathogen interaction | 0 | 0 | 0 | 34 | 24 | 27 |
| ko00360 | Phenylalanine metabolism | 0 | 0 | 0 | 31 | 0 | 0 |
| ko00520 | Amino sugar and nucleotide sugar metabolism | 0 | 3 | 0 | 23 | 20 | 18 |
| ko00940 | Phenylpropanoid biosynthesis | 5 | 5 | 0 | 10 | 29 | 32 |
| ko00941 | Flavonoid biosynthesis | 1 | 0 | 7 | 15 | 11 | 12 |
| ko00904 | Diterpenoid biosynthesis | 1 | 4 | 7 | 15 | 13 | 13 |
| ko00592 | Alpha-linolenic acid metabolism | 0 | 0 | 0 | 15 | 9 | 11 |
| ko00910 | Nitrogen metabolism | 1 | 2 | 0 | 12 | 10 | 10 |
| CK | T10-d10 | T20-d10 | T10-d20 | T20-d20 | |
|---|---|---|---|---|---|
| GA | |||||
| Bioactive GAs | |||||
| GA1 | 0.010 ± 0.002 d | 0.306 ± 0.048 b | 0.087 ± 0.061 cd | 0.511 ± 0.055 a | 0.183 ± 0.017 c |
| GA3 | 0.216 ± 0.018 | ND | ND | ND | ND |
| GA4 | 0.078 ± 0.013 b | 0.058 ± 0.011 b | 0.074 ± 0.011 b | 0.123 ± 0.028 a | 0.049 ± 0.010 b |
| GA7 | 0.153 ± 0.024 a | 0.054 ± 0.001 b | 0.041 ± 0.013 b | 0.057 ± 0.011 b | 0.033 ± 0.015 b |
| Precursors | |||||
| GA12 | ND | ND | ND | ND | ND |
| GA53 | 1.901 ± 0.258 a | 1.829 ± 0.241 a | 1.435 ± 0.163 b | 2.079 ± 0.073 a | 1.940 ± 0.145 a |
| GA20 | 0.038 ± 0.014 c | 0.051 ± 0.021 c | 0.068 ± 0.007 c | 0.271 ± 0.025 a | 0.164 ± 0.021 b |
| GA24 | 0.100 ± 0.029 b | 0.026 ± 0.005 c | 0.019 ± 0.008 c | 0.160 ± 0.014 a | 0.044 ± 0.017 c |
| GA9 | 0.093 ± 0.077 ab | 0.151 ± 0.028 a | 0.096 ± 0.052 ab | 0.052 ± 0.003 ab | 0.032 ± 0.006 b |
| Deactivated GAs | |||||
| GA8 | 0.178 ± 0.032 d | 0.965 ± 0.050 b | 1.087 ± 0.126 ab | 1.194 ± 0.098 a | 0.642 ± 0.018 c |
| GA29 | ND | ND | ND | 0.950 ± 0.240 | ND |
| GA51 | 1.020 ± 0.128 a | 0.501 ± 0.113 b | 0.436 ± 0.002 b | 0.244 ± 0.012 c | 0.071 ± 0.026 c |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Sun, M.; Xiang, H.; Liu, Y.; Li, H.; Qi, M.; Li, T. Low Overnight Temperature-Induced Gibberellin Accumulation Increases Locule Number in Tomato. Int. J. Mol. Sci. 2019, 20, 3042. https://doi.org/10.3390/ijms20123042
Li Y, Sun M, Xiang H, Liu Y, Li H, Qi M, Li T. Low Overnight Temperature-Induced Gibberellin Accumulation Increases Locule Number in Tomato. International Journal of Molecular Sciences. 2019; 20(12):3042. https://doi.org/10.3390/ijms20123042
Chicago/Turabian StyleLi, Yanbing, Meihua Sun, Hengzuo Xiang, Yudong Liu, Hui Li, Mingfang Qi, and Tianlai Li. 2019. "Low Overnight Temperature-Induced Gibberellin Accumulation Increases Locule Number in Tomato" International Journal of Molecular Sciences 20, no. 12: 3042. https://doi.org/10.3390/ijms20123042
APA StyleLi, Y., Sun, M., Xiang, H., Liu, Y., Li, H., Qi, M., & Li, T. (2019). Low Overnight Temperature-Induced Gibberellin Accumulation Increases Locule Number in Tomato. International Journal of Molecular Sciences, 20(12), 3042. https://doi.org/10.3390/ijms20123042