Expansion and Functional Diversification of SKP1-Like Genes in Wheat (Triticum aestivum L.)
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification of TaSKP1-Like Genes
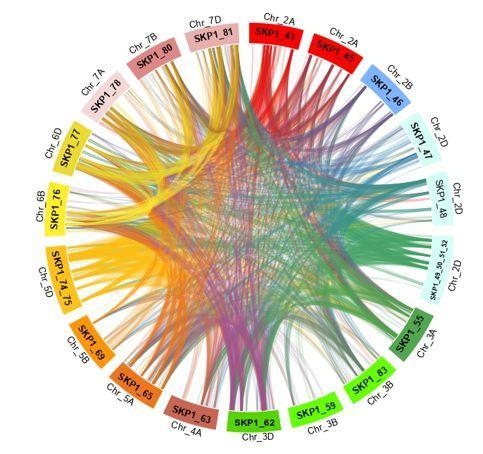
2.2. Chromosomal Location of TaSKP1-Like Gene
2.3. Phylogeny of the TaSKP1-Like Genes
2.4. Homoeologous Group Identification, Structure and Expression Profiles of TaSKP1-Like Genes
2.5. Duplications
2.6. Structural and Phylogenic Analysis of TaSKP1-Like Genes
2.7. Expression Profiles of the TaSKP1-Like Genes during Wheat Development
2.8. Expression Profiles of the TaSKP1-like Genes in Response to Different Abiotic Stresses in Leaves
2.9. Yeast Two-Hybrid (Y2H) Interaction between TaSKP1, FBX Proteins and Western Blots
3. Discussion
3.1. Expansion and Genomic Distribution of the TaSKP1-Like Gene Family
3.2. Fate of Duplicated Genes
3.3. Protein–Protein Interactions
4. Materials and Methods
4.1. Data Retrieval
4.2. Chromosomal Location of TaSKP1-Like Genes
4.3. Identification of Homoeologous Relationships between TaSKP1 Sequences
4.4. Duplications
4.5. Phylogeny of the TaSKP1-Like Genes
4.6. Meta-Analysis of the Expression of Wheat SKP1-Like Genes
4.7. Plant Materials and Growth Conditions
4.8. Total RNA Extraction and Isolation of TaSKP and TaFBX cDNAs
4.9. cDNA Cloning
4.10. Binary Yeast Two-Hybrid Analysis (Y2H)
4.10.1. Plasmid Constructs
4.10.2. Yeast Two-Hybrid (Y2H) Screening and Assays
4.11. Protein Extraction and Immunoblotting
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hershko, A.; Ciechanover, A. Mechanisms of intracellular protein breakdown. Annu. Rev. Biochem. 1982, 51, 335–364. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Genschik, P.; Marrocco, K.; Bach, L.; Noir, S.; Criqui, M.C. Selective protein degradation: A rheostat to modulate cell-cycle phase transitions. J. Exp. Bot. 2014, 65, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Wang, C.; Huang, J.; Yang, G.; Wu, C.; Zheng, C. SCF E3 ligase PP2-B11 plays a positive role in response to salt stress in Arabidopsis. J. Exp. Bot. 2015, 66, 4683–4697. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Callis, J. Ubiquitin ligases mediate growth and development by promoting protein death. Curr. Opin. Plant Biol. 2007, 10, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L. The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 2014, 5, 135. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Skaar, J.R.; Pagan, J.K.; Pagano, M. SnapShot: F-Box Proteins I. Cell 2009, 137, 1160–1161. [Google Scholar] [CrossRef]
- Feldman, R.M.R.; Correll, C.C.; Kaplan, K.B.; Deshaies, R.J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 1997, 91, 221–230. [Google Scholar] [CrossRef]
- Kamura, T.; Conrad, M.N.; Yan, Q.; Conaway, R.C.; Conaway, J.W. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999, 13, 2928–2933. [Google Scholar] [CrossRef]
- Gray, W.M.; del Pozo, J.C.; Walker, L.; Hobbie, L.; Risseeuw, E.; Banks, T.; Crosby, W.L.; Yang, M.; Ma, H.; Estelle, M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999, 13, 1678–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Liu, F.; Lechner, E.; Genschik, P.; Crosby, W.L.; Ma, H.; Peng, W.; Huang, D.; Xie, D. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 2002, 8, 1919–1935. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.; Santiago, F.E.; Jin, H.; Lin, D.; Schedl, T.; Kipreos, E.T. The Caenorhabditis elegans Skp1-related gene family: Diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol. 2002, 124, 277–287. [Google Scholar] [CrossRef]
- Guo, H.; Ecker, J.R. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2- dependent proteolysis of EIN3 transcription factor. Cell 2003, 115, 667–677. [Google Scholar] [CrossRef]
- Ni, W.; Xie, D.; Hobbie, L.; Feng, B.; Zhao, D.; Akkara, J.; Ma, H. Regulation of Flower Development by SCF Complexes1 in Arabidopsis. Plant Physiol. 2004, 134, 1574–1585. [Google Scholar] [CrossRef]
- Parry, G.; Estelle, M. Auxin receptors: A new role for F-box proteins. Curr. Opin. Cell Biol. 2006, 18, 152–156. [Google Scholar] [CrossRef]
- Porat, R.; Lu, P.; O’Neill, S.D. Arabidopsis SKP1, a homologue of a cell cycle regulator gene, is predominantly expressed in meristematic cells. Planta 1998, 204, 345–351. [Google Scholar] [CrossRef]
- Zhao, D.; Yang, M.; Solava, J.J.; Ma, H. The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev. Genet. 1999, 25, 209–223. [Google Scholar] [CrossRef]
- Zhao, D.; Yu, Q.; Chen, M.; Ma, H. The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis. Development 2001, 128, 2735–2746. [Google Scholar]
- Zhao, D.; Ni, W.; Feng, B.; Han, T.; Petrasek, M.G.; Ma, H. Members of the Arabidopsis-SKP1-like Gene Family Exhibit a Variety of Expression Patterns and May Play Diverse Roles in Arabidopsis. Plant Physiol. 2003, 133, 203–217. [Google Scholar] [CrossRef]
- Liu, F.; Ni, W.; Griffith, M.E.; Huang, Z.; Chang, C.; Peng, W.; Ma, H.; Xie, D. The ASK1 and ASK2 Genes Are Essential for Arabidopsis Early Development. Plant Cell 2004, 16, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Leebens-Mack, J.; Ni, W.; DePamphilis, C.W.; Ma, H. Highly Heterogeneous Rates of Evolution in the SKP1 Gene Family in Plants and Animals: Functional and Evolutionary Implications. Mol. Biol. Evol. 2004, 21, 117–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahloul, S.; Hajsalah El Beji, I.; Boulaflous, A.; Ferchichi, A.; Kong, H.; Mouzeyar, S.; Bouzidi, M.F. Structural, expression and interaction analysis of rice SKP1-like genes. DNA Res. 2013, 20, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Elzanati, O.; Roche, J.; Boulaflous-Stevens, A.; Mouzeyar, S.; Bouzidi, M.F. Genome-wide analysis, classification, expression and interaction of Physcomitrella patens SKP1-like (PpSKP) and F-box (FBX) genes. Plant Gene 2017, 12, 13–22. [Google Scholar] [CrossRef]
- Takahashi, N.; Kuroda, H.; Kuromori, T.; Hirayama, T.; Seki, M.; Shinozaki, K. Expression and Interaction Analysis of Arabidopsis Skp1-Related Genes. Plant Cell Physiol. 2004, 45, 83–91. [Google Scholar] [CrossRef] [Green Version]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; DePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. expVIP: A Customizable RNA-seq Data Analysis and Visualization Platform. Plant Physiol. 2016, 170, 2172–2186. [Google Scholar] [CrossRef]
- Clavijo, B.J.; Venturini, L.; Schudoma, C.; Accinelli, G.G.; Kaithakottil, G.; Wright, J. An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 2017, 27, 885–896. [Google Scholar] [CrossRef] [Green Version]
- Choulet, F.; Alberti, A.; Theil, S.; Glover, N.; Barbe, V.; Daron, J.; Pingault, L.; Sourdille, P.; Couloux, A.; Paux, E.; et al. Structural and functional partitioning of bread wheat chromosome 3B. Science 2014, 345, 1249721. [Google Scholar] [CrossRef]
- Smyth, G.K. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 1–25. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Farlow, A.; Meduri, E.; Schlöttereret, C. DNA double-strand break repair and the evolution of intron density. Trends Genet. 2011, 27, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wicker, T.; Buchmann, J.P.; Keller, B. Patching gaps in plant genomes results in gene movement and erosion of colinearity. Genome Res. Trends Genet. 2010, 20, 1229–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glover, N.M.; Daron, J.; Pingault, L.; Vandepoele, K.; Paux, E.; Feuillet, C. Small-scale gene duplications played a major role in the recent evolution of wheat chromosome 3B. Genome Biol. 2015, 16, 188. [Google Scholar] [CrossRef] [PubMed]
- Guérin, C.; Roche, J.; Allard, V.; Ravel, C.; Mouzeyar, S.; Bouzidi, M.F. Genome-wide analysis, expansion and expression of the NAC family under drought and heat stresses in bread wheat (T. Aestivum L.). PLoS ONE 2019, 14. [Google Scholar] [CrossRef] [PubMed]
- Schulman, B.A.; Carrano, A.C.; Jeffrey, P.D.; Bowen, Z.; Kinnucan, E.R.E.; Finnin, M.S.; Elledge, S.J.; Harper, J.W.; Pagano, M.; Pavletich, N.P. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 2000, 408, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Schulman, B.A.; Song, L.; Miller, J.J.; Jeffrey, P.D.; Wang, P.; Chu, C.; Koepp, D.M.; Elledge, S.J.; Paganok, M.; et al. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 2002, 416, 703–709. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [Green Version]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar]
- Risseeuw, E.P.; Daskalchuk, T.E.; Banks, T.W.; Liu, E.; Cotelesage, J.; Hellmann, H.; Estelle, M.; Somers, D.E.; Crosby, W.L. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 2003, 34, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M. UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Mulder, N.J.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Buillard, V.; Cerutti, L.; Copley, R.; et al. New developments in the InterPro database. Nucleic Acids Res. 2007, 35, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, M.; Kugler, K.G.; Sandve, S.R.; Zhan, B.; Rudi, H.; Hvidsten, T.R. International Wheat Genome Sequencing Consortium. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 2014, 345, 1250091. [Google Scholar] [CrossRef] [PubMed]
- Noé, L.; Kucherov, G. YASS: Enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 2005, 33, 540–543. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Rogozin, I.B.; Koonin, E.V.; Przytycka, T.M. Support for the Coelomata clade of animals from a rigorous analysis of the pattern of intron conservation. Mol. Biol. Evol. 2007, 24, 2583–2592. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2017, 33, 1870–1874. [Google Scholar] [CrossRef]
- Law, C.W.; Yunshun, C.; Shi, W.; Smyth, G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef] [Green Version]
- Bogorad, L.; Gubbins, E.J.; Krebbers, E.; Larrinua, I.M.; Mulligan, B.J. Cloning and physical mapping of maize plastid genes [Zea mays, corn]. Methods Enzymol. 1983, 97, 524–554. [Google Scholar]
- Printen, J.A.; Sprague, G.F. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 1994, 138, 609–619. [Google Scholar] [PubMed]








| Chromosome | A Subgenome | B Subgenome | D Subgenome |
|---|---|---|---|
| 1 | 1 | 1 | 2 |
| 2 | 3 | 2 | 7 |
| 3 | 3 | 9 | 8 |
| 4 | 3 | 2 | 2 |
| 5 | 7 | 7 | 10 |
| 6 | 3 | 4 | 4 |
| 7 | 5 | 5 | 4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
HajSalah El Beji, I.; Mouzeyar, S.; Bouzidi, M.-F.; Roche, J. Expansion and Functional Diversification of SKP1-Like Genes in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2019, 20, 3295. https://doi.org/10.3390/ijms20133295
HajSalah El Beji I, Mouzeyar S, Bouzidi M-F, Roche J. Expansion and Functional Diversification of SKP1-Like Genes in Wheat (Triticum aestivum L.). International Journal of Molecular Sciences. 2019; 20(13):3295. https://doi.org/10.3390/ijms20133295
Chicago/Turabian StyleHajSalah El Beji, Imen, Said Mouzeyar, Mohammed-Fouad Bouzidi, and Jane Roche. 2019. "Expansion and Functional Diversification of SKP1-Like Genes in Wheat (Triticum aestivum L.)" International Journal of Molecular Sciences 20, no. 13: 3295. https://doi.org/10.3390/ijms20133295
APA StyleHajSalah El Beji, I., Mouzeyar, S., Bouzidi, M. -F., & Roche, J. (2019). Expansion and Functional Diversification of SKP1-Like Genes in Wheat (Triticum aestivum L.). International Journal of Molecular Sciences, 20(13), 3295. https://doi.org/10.3390/ijms20133295