Molecular and Cellular Bases of Immunosenescence, Inflammation, and Cardiovascular Complications Mimicking “Inflammaging” in Patients with Systemic Lupus Erythematosus
Abstract
:1. Introduction
2. The Cellular Basis of Inflammaging in Patients with SLE
3. The Factors Contributing to Inflammaging and Cardiovascular Morbidities in Patients with SLE
4. Mitochondrial Dysfunction is One of the Crucial Up-Stream Factors in Inducing Oxidative Stresses and Immunosenescence in Patients with SLE
5. Defective Bioenergetics/Immunometabolism Allied with Mitochondrial Dysfunction Accelerates Metabolic Syndrome and Immunosenescence in Patients with SLE
6. Metabolomic Signatures Predisposing to the Metabolic Syndrome in Patients with SLE
7. Activation of AGE-RAGE System Induces Skin Autofluorescence, Cardiovascular Morbidity, and Pro-Inflammatory IL-6 Production in Patients with SLE
8. Molecular Basis of Oxidative and Nitrosative Stresses in Inducing Autoimmunity and Cardiovascular Morbidity in Patients with SLE
8.1. ROS-Induced Lipid Peroxidation and Histone Modifications Play a Role in Autoimmunity and Cardiovascular Morbidities in SLE
8.2. Role of Nitrosative Stress in Neoantigen Formation and Autoantibody Production in Patients with SLE
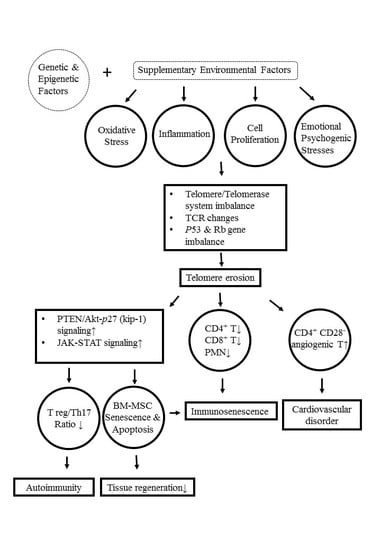
9. Molecular and Cellular Mechanisms Underlying Telomere/Telomerase Disequilibrium in Inducing Autoimmunity, Immunosenescence, and Bone Marrow-Derived Mesenchymal Stem Cell (BM-MSC) Senescence in Patients with SLE
10. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| AGE | advanced glycation end product |
| Akt | RAC-alpha serine/threonine-protein kinase |
| ANA | antinuclear antibodies |
| APL | antiphospholipid antibodies |
| ATA | anti-thyroglobulin antibodies |
| BM-MSC | bone marrow-derived mesenchymal stem cell |
| CD | cluster of differentiation |
| CD45RO | marker for memory T cell |
| CV | cardiovascular |
| DNA | deoxyribonucleic acid |
| EC | endothelial cell |
| fMLP | formyl-methionine-leucyl-phenylalanine |
| GC-MS | gas chromatography/mass spectrophotometry |
| GLUT | glucose transporter |
| GSH | reduced form glutathione |
| HMGB1 | high mobility group-box protein 1 |
| hOGG1 | 8-oxoguanine DNA glycosylase 1 |
| IFN | interferon |
| IL | interleukin |
| Inflammaging | inflammation inducing aging process |
| IP-10 | interferon-γ induced protein 10 |
| JAK-STAT | Janus kinase and signal transducer and activator of transcription |
| JUN | gene for ju-nana oncogene |
| kip-1 | cyclin-dependent kinase inhibitor 1 |
| LDL | low density lipoprotein |
| LPS | lipopolysaccharide |
| MAPK-ERK | mitogen-activated protein kinase- extracellular signal-regulated kinase |
| MCP-1 | monocyte chemoattractant protein 1 |
| MyD88 | myeloid differentiation primary response 88 |
| mTOR | mammalian target of rapamycin |
| mTORC | mTOR complex |
| mtDNA | mitochondrial DNA |
| MW | molecular weight |
| NET | neutrophil extracellular trap |
| NETosis | neutrophil apoptosis to form NET |
| NF-κB | nuclear factor of kappa-light-chain-enhancer of activated B cells |
| NGAL | neutrophil gelatinase-associated lipocalin |
| NMR | nuclear magnetic resonance |
| 8-OHdG | 8-hydroxy-2′-deoxyguanosine |
| PBMC | peripheral blood mononuclear cell |
| PMN | polymorphonuclear neutrophil |
| PTEN | phosphatase and tensin homolog |
| RAGE | receptor for AGE |
| RF | rheumatoid factor |
| RNS | reactive nitrogen species |
| ROS | reactive oxygen species |
| SLE | systemic lupus erythematosus |
| TCR | T cell receptor |
| Th | helper T cell |
| Treg | regulatory T cell |
References
- Kahlenberg, J.M.; Kaplan, M.J. The inflammasome and lupus-another innate immune mechanism contributing to disease pathogenesis? Curr. Opin. Rheumatol. 2014, 26, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Weidenbusch, M.; Kulkarni, O.; Anders, H.-J. The innate immune system in human systemic lupus erythematosus. Clin. Sci. 2017, 131, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-Y.; Li, K.-J.; Hsieh, S.-C.; Liao, H.-T.; Yu, C.-L. What’s wrong with neutrophils in lupus? Clin. Exp. Rheumatol. 2019, 37, 684–693. [Google Scholar] [PubMed]
- Zharkova, O.; Celhar, T.; Cravens, P.D.; Satherthwaite, A.B.; Fairhurst, A.-M.; Davis, L.S. Pathways leading to an immunological disease: Systemic lupus erythematosus. Rheumatology 2017, 56, i55–i66. [Google Scholar] [CrossRef] [PubMed]
- Morawski, P.A.; Bolland, S. Expanding the B cell centric view of systemic lupus erythematosus. Trends Immunol. 2017, 38, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Mohan, C.; Li, Q.-Z. Arraying autoantibodies in SLE-lessons learned. Curr. Mol. Med. 2015, 15, 456–461. [Google Scholar]
- Yaniv, G.; Twig, G.; Shor, D.B.-A.; Furer, A.; Sherer, Y.; Mozes, O.; Komisar, O.; Slonimsky, E.; Klang, E.; Lotan, E.; et al. A volcanic explosion of autoantibodies in systemic lupus erythematosus: A diversity of 180 different antibodies found in SLE patients. Autoimmunity Rev. 2015, 14, 75–79. [Google Scholar] [CrossRef]
- Montoya-Ortiz, G. Immunosenescence, aging and systemic lupus erythematosus. Autoimmune Dis. 2013, 2013, 1–5. [Google Scholar] [CrossRef]
- Fülöp, T., Jr.; Larbi, A.; Dupuis, G.; Pawelec, G. Ageing, autoimmunity and arthritis: Perturbations of TCR signal transduction pathways with ageing-a biochemical paradigm for the ageing immune system. Arthritis Res. Ther. 2003, 5, 290–302. [Google Scholar] [CrossRef]
- Ramos-Casals, M.; Garcia-Carrasco, M.; Brito, M.P.; López-Soto, A.; Font, J. Autoimmunity and geriatrics: Clinical significance of autoimmune manifestations in the elderly. Lupus 2003, 12, 341–355. [Google Scholar] [CrossRef]
- Watad, A.; Bragazzi, N.L.; Adawi, M.; Amital, H.; Toubi, E.; Porat, B.-S.; Shoenfeld, Y. Autoimmunity in the elderly: Insights from basic science and clinics-a mini-review. Gerontology 2017, 63, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Candore, G.; Caruso, C.; Jirillo, E.; Magrone, T.; Vasto, S. Low grade inflammation as a common pathogenetic denominator in age-related diseases: Novel drug targets for anti-ageing strategies and successful ageing achievement. Curr. Pharm. Des. 2010, 16, 584–596. [Google Scholar] [CrossRef] [PubMed]
- Fülöp, T.; Larbi, A.; Dupuis, G.; Le Page, A.; Frost, E.H.; Cohen, A.A.; Witkowski, J.M.; Franceschi, C. Immunosenescence and inflamm-aging as two sides of the same coin: Friends or foes? Front. Immunol. 2018, 8, 1960. [Google Scholar] [CrossRef] [PubMed]
- Fulop, T.; Larbi, A.; Douziech, N.; Fortin, C.; Guérard, K.-P.; Lesur, O.; Khalil, A.; Dupuis, G. Signal transduction and functional changes in neutrophils with aging. Aging Cell 2004, 3, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Wessels, I.; Jansen, J.; Rink, L.; Uciechowski, P. Immunosenescence of polymorphonuclear neutrophils. Sci. World J. 2010, 10, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Plowden, J.; Renshaw-Hoelscher, M.; Engleman, C.; Katz, J.; Sambhara, S. Innate immunity in aging: Impact on macrophage function. Aging Cell 2004, 3, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Solana, R.; Tarazona, R.; Gayoso, I.; Lesur, O.; Dupuis, G.; Fulop, T. Innate immunosenescence: Effect of aging on cells and receptors of the innate immune system in humans. Semin. Immunol. 2012, 24, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Hazeldine, J.; Lord, J.M. Innate immunosenescence: Underlying mechanisms and clinical relevance. Biogerontology 2015, 16, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Gounder, S.S.; Abdullah, B.J.J.; Radzuanb, N.E.I.B.M.; Zain, F.D.B.M.; Sait, N.B.M.; Chua, C.; Subramani, B. Effect of aging on NK cell population and their proliferation at ex vivo culture condition. Anal. Cell Pathol. (Amst.) 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Katsiari, C.G.; Liossie, S.N.; Sfikakis, P.P. The pathophysiologic role of monocytes and macrophages in systemic lupus erythematosus. Semin. Arthritis Rheum. 2010, 39, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Mackern-Oberti, J.P.; Llanos, C.; Riedel, C.A.; Bueno, S.M.; Kalergis, A.M. Contribution of dendritic cells to the autoimmune pathology of systemic lupus erythematosus. Immunology 2015, 146, 497–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hervier, B.; Beziat, V.; Haroche, J.; Mathian, A.; Lebon, P.; Ghillani-Dalbin, P.; Musset, L.; Debrè, P.; Amoura, Z.; Vieillard, V. Phenotype and function of natural killer cells in systemic lupus erythematosus: Excess interferon-γ production in patients with active disease. Arthritis Rheum. 2011, 63, 1698–1706. [Google Scholar] [CrossRef] [PubMed]
- Henriques, A.; Teixeira, L.; Inês, L.; Carvalheiro, T.; Gonçalves, A.; Martinho, A.; Pais, M.L.; da Silva, J.A.; Paiva, A. NK cells dysfunction in systemic lupus erythematosuas: Relation to disease activity. Clin. Rheumatol. 2013, 32, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Tsokos, G.C.; Lo, M.S.; Costa Reis, P.; Sullivan, K.E. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat. Rev. Rheumatol. 2016, 12, 716–730. [Google Scholar] [CrossRef] [PubMed]
- Rose, T.; Dörner, T. Drivers of the immunopathogenesis in systemic lupus erythematosus. Best Pract. Res. Clin. Rheumatol. 2017, 31, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.; Weber, C.; Storch, S.; Federlin, K. Relationship between CD5+ B lymphocytes and the activity of systemic autoimmunity. Clin. Immunol. Immunopathol. 1990, 56, 219–225. [Google Scholar] [CrossRef]
- Omar, H.H.; Nasef, S.I.; Omar, H.H.; Ghaly, M.S. CD5+ B lymphocytes in systemic lupus erythematosus patients: Relation to disease activity. Clin. Rheumatol. 2017, 36, 2719–2726. [Google Scholar] [CrossRef] [PubMed]
- Ghodke-Puranik, Y.; Niewold, T.B. Immunogenetics of systemic lupus erythematosus: A comprehensive review. J. Autoimmun. 2015, 64, 125–136. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, T.; Niewold, T.B. Genetics of human lupus nephritis. Clin. Immunol. 2017, 185, 32–39. [Google Scholar]
- Hiraki, L.T.; Silverman, E.D. Genomics of systemic lupus erythematosus: Insights gained by studying monogenic young-onset systemic lupus erythematosus. Rheum. Dis. Clin. North Am. 2017, 43, 415–434. [Google Scholar] [CrossRef]
- Goulielmos, G.N.; Zervou, M.I.; Vazgiourakis, V.M.; Ghodke-Puranik, Y.; Garyballos, A.; Niewold, T.B. The genetics and molecular pathogenesis of systemic lupus erythematosus (SLE) in populations of different ancestry. Gene 2018, 668, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Yin, H.; Wang, L.; Gershwin, M.E.; Lu, Q. The critical role of epigenetics in systemic lupus erythematosus and autoimmunity. J. Autoimmun. 2016, 74, 118–138. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, C.M. Epigenetics in SLE. Clin. Rheumatol. Rep. 2017, 19, 58. [Google Scholar] [CrossRef] [PubMed]
- Lai, N.-S.; Koo, M.; Yu, C.-L.; Lu, M.-C. Immunopathogenesis of systemic lupus erythematosus and rheumatoid arthritis: The role of aberrant expression of non-coding RNAs in T cells. Clin. Exp. Immunol. 2017, 187, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Zununi Vahed, S.; Nakhjavani, M.; Etemadi, J.; Jamshidi, H.; Jadidian, N.; Pourlak, T.; Abediazar, S. Altered levels of immune-regulatory microRNAs in plasma samples of patients with lupus nephritis. Bioimpacts 2018, 8, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Honarpisheh, M.; Köhler, P.; von Rauchhaupt, E.; Lech, M. The involvement of microRNAs in modulation of innate and adaptive immunity in systemic lupus erythematosus and lupus nephritis. J. Immunol. Res. 2018, 2018, 1–5. [Google Scholar] [CrossRef] [PubMed]
- McMurray, R.W. Sex-hormones in the pathogenesis of systemic lupus erythematosus. Front. Biosci. 2001, 6, E193–E206. [Google Scholar] [CrossRef]
- Assad, S.; Khan, H.H.; Ghazanfar, H.; Khan, Z.H.; Mansor, S.; Rahman, M.A.; Khan, G.H.; Zafar, B.; Tariq, U.; Malik, S.A. Role of sex hormone levels and psychological stress in the pathogenesis of autoimmune diseases. Cureus 2017, 9, e1315. [Google Scholar] [CrossRef]
- Sarzi-Puttini, P.; Atzeni, F.; Iaccarino, L.; Doria, A. Environment and systemic lupus erythematosus: An overview. Autoimmunity 2005, 38, 465–472. [Google Scholar] [CrossRef]
- Parks, C.G.; de Souza Espindola Santos, A.; Barbhaiya, M.; Costenbader, K.H. Understanding the role of environmental factors in the development of systemic lupus erythematosus. Best Pract. Res. Clin. Rheumatol. 2017, 31, 306–320. [Google Scholar] [CrossRef]
- Marion, T.N.; Postlethwaite, A.E. Chance, genetics, and the heterogeneity of disease and pathogenesis in systemic lupus erythematosus. Semin. Immunopathol. 2014, 36, 495–517. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-T.; Lin, C.-S.; Chen, W.-S.; Liao, H.-T.; Tsai, C.-Y.; Wei, Y.-H. Leukocyte mitochondrial DNA alteration in systemic lupus erythematosus and its relevance to the susceptibility to lupus nephritis. Int. J. Mol. Sci. 2012, 13, 8853–8868. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-T.; Wu, T.-H.; Lin, C.-S.; Lee, C.S.; Wei, Y.-H.; Tsai, C.-Y.; Chang, D.-M. The pathogenesis of systemic lupus erythematosus-from the viewpoint of oxidative stress and mitochondrial dysfunction. Mitochondrion 2016, 30, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Leishangthem, B.D.; Sharma, A.; Bhatnagar, A. Role of altered mitochondria functions in the pathogenesis of systemic lupus erythematosus. Lupus 2016, 25, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-T.; Wu, T.-H.; Lin, C.-S.; Lee, C.-S.; Pan, S.-C.; Chang, D.-M.; Wei, Y.H.; Tsai, C.-Y. Oxidative DNA and mitochondrial DNA change in patients with SLE. Front. Biosci. (Landmark Ed.) 2017, 22, 493–503. [Google Scholar] [PubMed] [Green Version]
- Yang, S.K.; Zhang, H.R.; Shi, S.P.; Zhu, Y.Q.; Song, N.; Dai, Q.; Zhang, W.; Gui, M.; Zhang, H. The role of mitochondria in systemic lupus erythematosus: A glimpse of various pathogenetic mechanisms. Curr. Medicin. Chem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Li, K.-J.; Wu, C.-H.; Hsieh, S.-C.; Lu, M.-C.; Tsai, C.-Y.; Yu, C.-L. Deranged bioenergetics and defective redox capacity in T lymphocytes and neutrophils are related to cellular dysfunction and increased oxidative stress in patients with active systemic lupus erythematosus. Clin. Dev. Immunol. 2012, 4. [Google Scholar] [CrossRef]
- Morel, L. Immunometabolism in systemic lupus erythematosus. Nat. Rev. Rheumatol. 2017, 13, 280–290. [Google Scholar] [CrossRef]
- Vlassopoulos, A.; Lean, M.E.J.; Combet, E. Oxidative stress, protein glycation and nutrition-interactions relevant to health and disease throughout the lifecycle. Proc. Nutr. Soc. 2014, 73, 430–438. [Google Scholar] [CrossRef]
- Kurien, B.T.; Scofield, R.H. Lipid peroxidation in systemic lupus erythematosus. Indian J. Exp. Biol. 2006, 44, 349–356. [Google Scholar]
- McGuire, P.J. Mitochondrial dysfunction and the aging immune system. Biology 2019, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Schleicher, E.; Friess, U. Oxidative stress, AGE, and atherosclerosis. Kidney Int. Suppl. 2007, 106, S17–S26. [Google Scholar] [CrossRef] [PubMed]
- Rus, V.; Via, C.S. Telomeres, telomerase and lupus: The long and short of it. Clin. Immunol. 2001, 99, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Kurosaka, D.; Yasuda, J.; Yoshida, K.; Yoneda, A.; Yasuda, C.; Kingetsu, I.; Toyokawa, Y.; Yokoyama, T.; Saito, S.; Yamada, A. Abnormal telomerase activity and telomere length in T and B cells from patients with systemic lupus erythematosus. J. Rheumatol. 2006, 33, 1102–1107. [Google Scholar] [PubMed]
- Georgin-Lavialle, S.; Aouba, A.; Mouthon, L.; Londono-Vallejo, J.A.; Lepelletier, Y.; Gabet, A.-S.; Hermine, O. The telomere/telomerase system in autoimmune and systemic immune-medidated diseases. Autoimmun. Rev. 2010, 9, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Fathollahi, A.; Gabalou, N.B.; Aslani, S. Mesenchymal stem cell transplantation in systemic lupus erythematosus, a mesenchymal stem cell disorder. Lupus 2018, 27, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Cannizzo, E.S.; Clement, C.C.; Sahu, R.; Follo, C.; Santambrogio, L. Oxidative stress, inflamm-aging and immunosenescence. J. Proteomics 2011, 74, 2313–2323. [Google Scholar] [CrossRef]
- Cevenini, E.; Monti, D.; Franceschi, C. Inflamm-ageing. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 14–20. [Google Scholar] [CrossRef]
- Larbi, A.; Fülöp, T.; Pawelec, G. Immune receptor signaling, aging and autoimmunity. Adv. Exp. Med. Biol. 2008, 640, 312–324. [Google Scholar]
- Bulati, M.; Caruso, C.; Colonna-Romano, G. From lymphopoiesis to plasma cells differentiation. The age-related modifications of B cell compartment are influenced by “inflamm-ageing”. Ageing Res. Rev. 2017, 36, 125–136. [Google Scholar] [CrossRef]
- López, P.; Rodríguez-Carrio, J.; Martinez-Zapico, A.; Caminal-Montero, L.; Suarez, A. Senescent profile of angiogenic T cells from systemic lupus erythematosus patients. J. Leukoc. Biol. 2016, 99, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Bella, S.D.; Mavilio, D. Senescent angiogenic T cells: The use of CD28 makes the difference in endothelial homeostasis. J. Leukoc. Biol. 2016, 99, 399–401. [Google Scholar] [CrossRef] [PubMed]
- Thewissen, M.; Somers, V.; Venken, K.; Linsen, L.; van Paassen, P.; Geusens, P.; Damoiseaux, J.; Stinissen, P. Analyses of immunosenescent markers in patients with autoimmune disease. Clin. Immunol. 2007, 123, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.X.; Fu, C.W.; Jiang, F.; Ye, L.X.; Meng, W. Association of the interleukin-6 polymorphisms with systemic lupus erythematosus: A meta-analysis. Lupus 2015, 24, 1308–1317. [Google Scholar] [CrossRef] [PubMed]
- Rekik, R.; Smiti Khanfir, M.; Larbi, T.; Zamali, I.; Beldi-Ferchiou, A.; Kammoun, O.; Marzouki, S.; Hamzaoui, S.; Mrad, S.; Barbouche, M.R.; et al. Impaired TGF-β signaling in patients with active systemic lupus erythematosus is associated with an overexpression of IL-22. Cytokine 2018, 108, 182–189. [Google Scholar] [CrossRef]
- Van den Hoogen, L.L.; Sims, G.P.; van Roon, J.A.; Fritsch-Stork, R.D. Aging and systemic lupus erythematosus-immunosenescence and beyond. Curr. Aging Sci. 2015, 8, 158–177. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Zhang, X.; Zheng, S.; Khanabdali, R.; Kalionis, B.; Wu, J.; Wan, W.; Tai, X. An update on inflamm-aging: Mechanisms, prevention, and treatment. J. Immunol. Res. 2016, 2016, 1–12. [Google Scholar] [CrossRef]
- Garrido, A.; Cruces, J.; Ceprian, N.; Vara, E.; de la Fuente, M. Oxidative- inflammatory stress in immune cells from adult mice with premature aging. Int. J. Mol. Sci. 2019, 20, E769. [Google Scholar] [CrossRef]
- Peri, A. Oxidative stress in the pathology and treatment of systemic lupus erythematosus. Nat. Rev. Rheumatol. 2013, 9, 674–686. [Google Scholar] [Green Version]
- Shah, D.; Mahajan, N.; Sah, S.; Nath, S.K.; Paudyal, B. Oxidative stress and its biomarkers in systemic lupus erythematosus. J. Biomed. Sci. 2014, 21, 23. [Google Scholar] [CrossRef]
- Bauer, M.E.; De la Fuente, M. The role of oxidative and inflammatory stress and persistent viral infections in immunosenescence. Mech. Ageing Dev. 2016, 158, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Gergely, P., Jr.; Grossman, C.; Niland, B.; Puskas, F.; Neupane, H.; Allam, F.; Banki, K.; Phillips, P.E.; Perl, A. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum. 2002, 46, 175–190. [Google Scholar] [CrossRef]
- Perl, A.; Gergely, P., Jr.; Banki, K. Mitochondrial dysfunction in T cells of patients with systemic lupus erythematosus. Int. Rev. Immunol. 2004, 23, 293–313. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, D.; Perl, A. Metabolic control of T cell activation and death in SLE. Autoimmun. Rev. 2009, 8, 184–189. [Google Scholar] [CrossRef] [Green Version]
- Perl, A.; Hanczko, R.; Doherty, E. Assessment of mitochondrial dysfunction in lymphocytes of patients with systemic lupus erythematosus. Methods Mol. Biol. 2012, 900, 61–89. [Google Scholar]
- López-López, L.; Nieves-Plaza, M.; del R Castro, M.; Font, Y.M.; Torres-Ramos, C.; Vilá, L.M.; Ayala-Peňa, S. Mitochondrial DNA damage is associated with damage accrual and disease duration in patients with systemic lupus erythematosus. Lupus 2014, 23, 1133–1141. [Google Scholar]
- Vyshkina, T.; Sylvester, A.; Sadiq, S.; Bonilla, E.; Canter, J.A.; Perl, A.; Kalman, B. Association of common mitochondrial DNA variants with multiple sclerosis and systemic lupus erythematosus. Clin. Immunol. 2008, 129, 31–35. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.-J.; Cheng, T.-T.; Chen, C.-J.; Chang, W.-N.; Tsai, N.-W.; Kung, C.T.; Wang, H.-C.; Lin, W.-C.; Huang, C.-C.; Chang, Y.-T.; et al. Investigation of the caspase-dependent mitochondrial apoptotic pathway in mononuclear cells of patients with systemic lupus erythematosus. J. Transl. Med. 2014, 12, 303. [Google Scholar] [CrossRef]
- Lee, H.-T.; Lin, C.-S.; Lee, C.-S.; Tsai, C.-Y.; Wei, Y.-H. Increased 8-hydroxy-2′-deoxyguanosine in plasma and decreased mRNA expression of human 8-oxoguanine DNA glycosylase 1, anti-oxidant enzymes, mitochondrial biogenesis-related proteins and glycolytic enzymes in leucocytes in patients with systemic lupus erythematosus. Clin. Exp. Immunol. 2014, 176, 66–77. [Google Scholar]
- Lee, H.T.; Lin, C.S.; Lee, C.S.; Tsai, C.Y.; Wei, Y.H. The role of hOGG1 C1245G polymorphism in susceptibility to lupus nephritis and modulation of the plasma 8-OHdG in patients with systemic lupus erythematosus. Int. J. Mol. Sci. 2015, 16, 3757–3768. [Google Scholar] [CrossRef]
- Yang, C.-C.; Hsieh, S.-C.; Li, K.-J.; Wu, C.-H.; Lu, M.-C.; Tsai, C.-Y.; Yu, C.-L. Urinary neutrophil gelatinase -associated lipocalin is a potential biomarker for renal damage in patients with systemic lupus erythematosus. J. Biomed. Biotechnol. 2012, 8, 759313. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-T.; Lin, C.-S.; Pan, S.-C.; Wu, T.-H.; Lee, C.-S.; Chang, D.-M.; Tsai, C.-Y.; Wei, Y.-H. Alterations of oxygen consumption and extracellular acidification rates by glutamine in PBMCs of SLE patients. Mitochondrion 2019, 44, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, Y.; Iwasaki, Y.; Okamura, T.; Fujio, K.; Yamamoto, K. The metabolic regulation in immune cells and pathogenesis of systemic lupus eryt]hematosus-toward new therapeutic applications. Nihon Rinsho Meneki Gakkai Kaishi 2017, 40, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.-Y.; Mauro, C. Similarities in the metabolic reprogramming of immune system and endothelium. Front. Immunol. 2017, 8, 837. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Matteson, E.L.; Goronzy, J.J.; Weyand, C.M. T-cell metabolism in autoimmune disease. Arthritis Res. Ther. 2015, 17, 29. [Google Scholar] [CrossRef] [PubMed]
- Lightfoot, Y.L.; Blanco, L.P.; Kaplan, M.J. Metabolic abnormalities and oxidative stress in lupus. Curr. Opin. Rheumatol. 2017, 29, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, Y.; Iwasaki, Y.; Fujio, K.; Yamamoto, K. Metabolism as a key regulator in the pathogenesis of systemic lupus erythematosus. Sem. Arthritis Rheum. 2019, 48, 1142–1145. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.C.; Titov, A.A.; Sivakumar, R.; Li, W.; Morel, L. Immune cell metabolism in systemic lupus erythematosus. Curr. Rheumatol. Rep. 2016, 18, 66. [Google Scholar] [CrossRef]
- Rhoads, J.P.; Major, A.S.; Rathmell, J.C. Fine tuning of immunometabolism for the treatment of rheumatic diseases. Nat. Rev. Rheumatol. 2017, 13, 313–320. [Google Scholar] [CrossRef]
- Ugarte-Gil, M.F.; Sánchez-Zúňiga, C.; Gamboa-Cárdenas, R.V.; Aliaga-Zamudio, M.; Zevallos, F.; Tineo-Pozo, G.; Cucho-Venegas, J.M.; Mosqueira-Riveros, A.; Perich-Campos, R.A.; Alfaro-Lozano, J.L.; et al. Circulating naïve and memory CD4+ T cells and metabolic syndrome in patients with systemic lupus erythematosus data from a primarily Mestizo population. Rheumatology 2015, 54, 1302–1307. [Google Scholar] [CrossRef]
- Perl, A. Activation of mTOR (mechanistic target of rapamycin) in rheumatic diseases. Nat. Rev. Rheumatol. 2016, 12, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Femandez, D.; Perl, A. mTOR signaling: A central pathway to pathogenesis in systemic lupus erythematosus? Discov. Med. 2010, 9, 173–178. [Google Scholar]
- Perl, A. mTOR activation is a biomarker and a central pathway to autoimmune disorders, cancer, obesity, and aging. Ann. N.Y. Acad. Sci. 2015, 1346, 33–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, N.; Perl, A. Metabolism as a target for modulation in autoimmune diseases. Trends Immunol. 2018, 39, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.-W.; Hanczko, R.; Bonilla, E.; Caza, T.N.; Clair, B.; Bartos, A.; Miklossy, G.; Jimah, J.; Doherty, E.; Tily, H.; et al. N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: A randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 2012, 64, 2937–2946. [Google Scholar] [CrossRef] [PubMed]
- Perl, A.; Hanczko, R.; Lai, Z.-W.; Oaks, Z.; Kelly, R.; Borsuk, R.; Asara, J.M.; Phillips, P.E. Comprehensive metabolome analyses reveal N-acetylcysteine-responsive accumulation of Kynurenine in systemic lupus erythematosus: Implications for activation of the mechanistic target of rapamycin. Metabolomics 2015, 11, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.-W.; Kelly, R.; Winans, T.; Marchena, I.; Shadakshari, A.; Yu, J.; Dawood, M.; Garcia, R.; Tily, H.; Francis, L.; et al. Sirolimus in patients with clinically active systemic lupus erythematosus resistant to, or intolerant of, conventional medications: A single-arm, open-label, phase 1/2 trial. Lancet 2018, 391, 1186–1196. [Google Scholar] [CrossRef]
- Duval, N.; Vacano, G.N.; Patterson, D. Rapamycin treatment ameliorates age-related accumulation of toxic metabolic intermediates in brains of the Ts65Dn mouse model of Down syndrome and aging. Front. Aging Neurosci. 2018, 10, 263. [Google Scholar] [CrossRef]
- Li, W.; Sivakumar, R.; Titov, A.A.; Choi, S.-C.; Morel, L. Metabolic factors that contribute to lupus pathogenesis. Crit. Rev. Immunol. 2016, 36, 75–98. [Google Scholar] [CrossRef]
- Ouyang, X.; Dai, Y.; Wen, J.L.; Wang, L.X. 1H NMR-based metabolomic study of metabolic profiling for systemic lupus erythematosus. Lupus 2011, 20, 1411–1420. [Google Scholar] [CrossRef]
- Ding, H.; Mohan, C. Connective tissue diseases: Promises and challenges of metabolomics in SLE. Nat. Rev. Rheumatol. 2016, 12, 627–628. [Google Scholar] [CrossRef] [PubMed]
- Guma, M.; Tiziani, S.; Firestein, G.S. Metabolomics in rheumatic diseases: Desperately seeking biomarkers. Nat. Rev. Rheumatol. 2016, 12, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Teruel, M.; Chamberlain, C.; Alarcón-Riquelme, M.E. Omics studies: Their use in diagnosis and reclassification of SLE and other systemic autoimmune diseases. Rheumatology 2017, 56 (Suppl. 1), i78–i87. [Google Scholar] [CrossRef] [PubMed]
- Davi, G.; Falco, A. Oxidant stress, inflammation and atherogenesis. Lupus 2005, 14, 760–764. [Google Scholar] [CrossRef] [PubMed]
- De Leeuw, K.; Graaff, R.; de Vries, R.; Dullaart, R.P.; Smit, A.J.; Kallenberg, C.G.; Bijl, M. Accumulation of advanced glycation endproducts in patients with systemic lupus erythematosus. Rheumatology 2007, 46, 1551–1556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nienhuis, H.L.; de Leeuw, K.; Bijzet, J.; Smit, A.; Schalkwijk, C.G.; Graaff, R.; Kallenberg, C.G.; Bijl, M. Skin autofluorescence is increased in systemic lupus erythematosus but is not reflected by elevated plasma levels of advanced glycation endproducts. Rheumatology 2008, 47, 1554–1558. [Google Scholar] [CrossRef] [Green Version]
- Meerwaldt, R.; Hartog, J.W.L.; Graaff, R.; Huisman, R.J.; Links, T.P.; den Hollander, N.C.; Thorpe, S.R.; Baynes, J.W.; Navis, G.; Gans, R.O.B.; et al. Skin autofluorescence, a measure of cumulative metabolic stress and advanced glycation endproducts, predicts mortality in hemodialysis patients. J. Am. Soc. Nephrol. 2005, 16, 3687–3693. [Google Scholar] [CrossRef]
- Sims, G.P.; Rowe, D.C.; Rietdijk, S.T.; Herbst, R.; Coyle, A.J. HMGB1 and RAGE in inflammation and cancer. Annu. Rev. Immunol. 2010, 28, 367–388. [Google Scholar] [CrossRef]
- Pan, H.F.; Wu, G.C.; Li, W.P.; Li, X.P.; Ye, D.Q. High mobility group box 1: A potential therapeutic target for systemic lupus erythematosus. Mol. Biol. Rep. 2010, 37, 1191–1195. [Google Scholar] [CrossRef]
- Ma, C.Y.; Ma, J.-L.; Jiao, Y.L.; Li, J.F.; Wang, L.C.; Yang, Q.R.; You, L.; Cui, B.; Chen, Z.J.; Zhao, Y.R. The plasma level of soluble receptor for advanced glycation end products is decreased in patients with systemic lupus erythematosus. Scand. J. Immunol. 2012, 75, 614–622. [Google Scholar] [CrossRef]
- Yu, S.L.; Wong, C.K.; Szeto, C.C.; Li, E.K.; Cai, Z.; Tam, L.S. Members of the receptor for advanced glycation end products axis as potential therapeutic targets in patients with lupus nephritis. Lupus 2015, 24, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Martens, H.A.; Nienhuis, H.L.A.; Gross, S.; van der Steege, G.; Brouwer, E.; Berden, J.H.M.; de Sévaux, R.G.L.; Derksen, R.H.W.M.; Voskuyl, A.E.; Berger, S.P.; et al. Receptor for advanced glycation end products (RAGE) polymorphisms are associated with systemic lupus erythematosus and disease severity in lupus nephritis. Lupus 2012, 21, 959–968. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.-Y.; Wu, C.-H.; Lu, C.-H.; Kuo, Y.-M.; Li, K.-J.; Hsieh, S.-C.; Yu, C.-L. Advanced glycation end products of bovine serum albumin suppressed Th1/Th2 cytokine but enhanced monocyte IL-6 gene expression via MAPK-ERK and MyD88 transduced NF-κB p50 signaling pathways. Molecules 2019, 24, E2461. [Google Scholar] [CrossRef] [PubMed]
- Ryan, B.J.; Nissim, A.; Winyard, P.G. Oxidative post-translational modifications and their involvement in the pathogenesis of autoimmune diseases. Redox Biol. 2014, 2, 715–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Shobaili, H.A.; Rasheed, Z. Physicochemical and immunological studies on mitochondrial DNA modified by peroxynitrite: Implications of neo-epitopes of mitochondrial DNA in the etiopathogenesis of systemic lupus erythematosus. Lupus 2013, 22, 1024–1037. [Google Scholar] [CrossRef] [PubMed]
- Kurien, B.T.; Hensley, K.; Bachmann, M.; Scofield, R.H. Oxidatively modified autoantigens in autoimmune diseases. Free Radic. Biol. Med. 2006, 41, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Mir, A.R.; Moinuddin. Glycoxidation of histone proteins in autoimmune disorders. Clin. Chim. Acta 2015, 450, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yang, X.; Zou, H.; Li, M. Oxidative stress and Treg and Th17 dysfunction in systemic lupus erythematosus. Oxidative Med. Cell. Longevity 2016, 1, 1–9. [Google Scholar] [CrossRef]
- Chirino, Y.I.; Orozco-Ibarra, M.; Pedraza-Chaverri, J. Role of peroxynitrite anion in different diseases. Rev. Investig. Clin. 2006, 58, 350–358. [Google Scholar]
- Ahmad, R.; Rasheed, Z.; Ahsan, H. Biochemical and cellular toxicology of peroxyritrite: Implications in cell death and autoimmune phenomenon. Immunopharmacol. Immunotoxicol. 2009, 31, 388–396. [Google Scholar] [CrossRef]
- Ahsan, H. 3-nitrotyrosine: A biomarker of nitrogen free radical species modified proteins in systemic autoimmune conditions. Human Immunol. 2013, 74, 1392–1399. [Google Scholar] [CrossRef] [PubMed]
- Bartesaghi, S.; Radi, R. Fundamentals on the biochemistry of peroxynitrite and protein tyrosine nitration. Redox Biol. 2018, 14, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Ahsan, H. Role of peroxynitrite-modified biomolecules in the etiopathogenesis of systemic lupus erythematosus. Clin. Exp. Med. 2014, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Alam, K.; Zafaryab, M.; Rizvi, M.M.A. Peroxynitrite-modified histone as a pathophysiological biomarker in autoimmune diseases. Biochimie 2017, 140, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Arif, Z.; Neelofar, K.; Tarannum, A.; Arfat, M.Y.; Ahmad, S.; Zaman, A.; Khan, M.A.; Badar, A.; Islam, S.N.; Iqubal, M.A. SLE autoantibodies are well recognized by peroxynitrite-modified-HSA: Its implications in the pathogenesis of SLE. Int. J. Biol. Macromol. 2018, 106, 1240–1249. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Hussain, A.; Ahsan, H. Peroxynitrite: Cellular pathology and implications in autoimmunity. J. Immunoassay Immunochem. 2019, 40, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Zakian, V.A. Telomeres: Beginning to understand the end. Science 1995, 270, 1601–1607. [Google Scholar] [CrossRef] [PubMed]
- Greider, C.W. Telomeres do D-loop-T-loop. Cell 1999, 97, 419–422. [Google Scholar] [CrossRef]
- Hodes, R.J. Telomere length, aging and somatic cell turnover. J. Exp. Med. 1999, 190, 153–156. [Google Scholar] [CrossRef]
- Weng, N.P.; Levine, B.L.; June, C.H.; Hodes, R.J. Regulated expression of telomerase activity in human T lymphocyte development and activation. J. Exp. Med. 1996, 183, 2471–2479. [Google Scholar] [CrossRef]
- Honda, M.; Mengesha, E.; Albano, S.; Nichols, W.S.; Wallace, D.J.; Metzger, A.; Klinenberg, J.R.; Linker-Israeli, M. Telomere shortening and decreased replicative potential, contrasted by continued proliferation of telomerase-positive CD8+CD28lo T cells in patients with systemic lupus erythematosus. Clin. Immunol. 2001, 99, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Katayama, Y.; Kohriyama, K. Telomerase activity in peripheral mononuclear cells of systemic connective tissue diseases. J. Rheumatol. 2001, 28, 288–291. [Google Scholar] [PubMed]
- Kurosaka, D.; Yasuda, J.; Yoshida, K.; Yokoyama, T.; Ozawa, Y.; Obayashi, Y.; Kingetsu, I.; Saito, S.; Yamada, A. Telomerase activity and telomere length of peripheral blood mononuclear cells in SLE patients. Lupus 2003, 12, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Kurosaka, D. Abnormalities in lymphocyte telomerase activity and telomere length in systemic lupus erythematosus. Nihon Rinsho Meneki Gakkai Kaishi 2007, 30, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Georgin-Lavialle, S.; Aouba, A.; Lepelletier, Y.; Gabet, A.S.; Hermine, O. Telomeres and telomerase: Relevance and future prospects in systemic lupus erythematosus. Rev. Med. Interne. 2010, 31, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Hsieh, S.-C.; Li, K.-J.; Lu, M.-C.; Yu, C.-L. Premature telomere shortening in polymorphonuclear neutrophils from patients with systemic lupus erythematosus is related to the lupus disease activity. Lupus 2007, 16, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.G.; Qing, Y.F.; Yang, Q.B.; Xie, W.G.; Zhao, M.C. Changes in the expression of telomere maintenance genes might play a role in the pathogenesis of systemic lupus erythematosus. Lupus 2011, 20, 820–828. [Google Scholar] [CrossRef]
- De Punder, K.; Heim, C.; Wadhwa, P.D.; Entringer, S. Stress and immunosenescence: The role of telomerase. Psychoneuroendocrinology 2019, 101, 87–100. [Google Scholar] [CrossRef]
- Tan, W.; Gu, Z.; Shen, B.; Jiang, J.; Meng, Y.; Da, Z.; Liu, H.; Tao, T.; Cheng, C. PTEN/Akt-p27(kip-1) signaling promotes the BM-MSCs senescence and apoptosis in SLE patients. J. Cell Biochem. 2015, 116, 1583–1594. [Google Scholar] [CrossRef]






| Parameters | Physiological Senescence | SLE | |
|---|---|---|---|
| • Immunological Functions | |||
| Neutrophil: | |||
| Phagocytosis | ↓ [8,14,15] | ↓ [3] | |
| Chemotactic capacity | ↓ | ↓ | |
| Response to bacterial products (fMLP, LPS) | ↓ | ↓ | |
| Response to IL-8 | ↓ | ↓ | |
| NETosis | ND | ↑ | |
| Link to Th1/Th2 cytokine ratio | ND | ↓ | |
| Macrophage/dendritic cell: | ↑ | ||
| Phagocytosis | ↓ [8,16,17,18] | ↓ [2,21,22] | |
| Chemotactic capacity | ↑ | ↑ | |
| Ability to stimulate lymphocyte | ↑ | ↑ | |
| Pro-inflammatory cytokine production | ↑ | ↑ | |
| Natural killer cell: | |||
| Cytotoxicity | ↓ [19] | ↓ [20,23,24] | |
| Proliferation | ↓ | ↓ | |
| T lymphocyte: | |||
| Th1 | ↓ [8,19] | ↓ [25,26] | |
| Th2 | ↑ | ↑ | |
| Th17 | ↑ | ↑ | |
| Treg | ↓ | ↓ | |
| CD4+CD 28null angiogenic T | ↑ | ↑ | |
| CD45RO+ T (memory T) | ↑ | ↑ | |
| B lymphocyte: | |||
| CD5+ B | ↑ [8,19] | ↑ [26,27] | |
| Hypergammaglobulinemia | ↑ | ↑ | |
| ANA | ↑ | ↑ | |
| RFs | ↑ | ↑ | |
| APL | ↑ | ↑ | |
| ATA | ↑ | ↑ | |
| • Common clinical features: | |||
| Infection rate | ↑ [9,10,11,12,13] | ↑ [2,3] | |
| Tumor incidence | ↑ | ↑ | |
| Cardiovascular diseases | ↑ | ↑ | |
| Cellular Basis of Inflammaging | Pathophysiological Effects |
|---|---|
| • Decrease in the expression and function of TCR and its co-receptors for antigens in T cells [59] | Susceptible to infections |
| • Decrease in circulating B cells due to reduction of new B cell migration from bone marrow and B lymphopoiesis [60] | Antibody production ↓ |
| •Shift from naïve to memory B cell [60] (naïve/memory B cell ratio ↓) | High affinity protective antibody production ↓ |
| • Impaired ability of memory B cell differentiation to plasma cells [60] | Antibody production ↓ |
| • CD4(+)CD28(+) angiogenic T cell ↓ [61,62,63] CD4(+)CD28(−) angiogenic T cell ↑ | Endothelial cell damage ↑ Cardiovascular morbidity ↑ |
| • Impaired IL-6/TGF-β balance [64,65] | Autoimmunity ↑, IL-22 ↑ |
| • Th17 cell ↑ | Inflammation ↑ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, C.-Y.; Shen, C.-Y.; Liao, H.-T.; Li, K.-J.; Lee, H.-T.; Lu, C.-S.; Wu, C.-H.; Kuo, Y.-M.; Hsieh, S.-C.; Yu, C.-L. Molecular and Cellular Bases of Immunosenescence, Inflammation, and Cardiovascular Complications Mimicking “Inflammaging” in Patients with Systemic Lupus Erythematosus. Int. J. Mol. Sci. 2019, 20, 3878. https://doi.org/10.3390/ijms20163878
Tsai C-Y, Shen C-Y, Liao H-T, Li K-J, Lee H-T, Lu C-S, Wu C-H, Kuo Y-M, Hsieh S-C, Yu C-L. Molecular and Cellular Bases of Immunosenescence, Inflammation, and Cardiovascular Complications Mimicking “Inflammaging” in Patients with Systemic Lupus Erythematosus. International Journal of Molecular Sciences. 2019; 20(16):3878. https://doi.org/10.3390/ijms20163878
Chicago/Turabian StyleTsai, Chang-Youh, Chieh-Yu Shen, Hsien-Tzung Liao, Ko-Jen Li, Hui-Ting Lee, Cheng-Shiun Lu, Cheng-Han Wu, Yu-Min Kuo, Song-Chou Hsieh, and Chia-Li Yu. 2019. "Molecular and Cellular Bases of Immunosenescence, Inflammation, and Cardiovascular Complications Mimicking “Inflammaging” in Patients with Systemic Lupus Erythematosus" International Journal of Molecular Sciences 20, no. 16: 3878. https://doi.org/10.3390/ijms20163878
APA StyleTsai, C. -Y., Shen, C. -Y., Liao, H. -T., Li, K. -J., Lee, H. -T., Lu, C. -S., Wu, C. -H., Kuo, Y. -M., Hsieh, S. -C., & Yu, C. -L. (2019). Molecular and Cellular Bases of Immunosenescence, Inflammation, and Cardiovascular Complications Mimicking “Inflammaging” in Patients with Systemic Lupus Erythematosus. International Journal of Molecular Sciences, 20(16), 3878. https://doi.org/10.3390/ijms20163878