Multitasking Rab Proteins in Autophagy and Membrane Trafficking: A Focus on Rab33b
Abstract
:1. Rab Proteins as Molecular Coordinators of Membrane Trafficking
2. Brief Overview of Autophagosome Formation
3. Regulation of Autophagy by Rab Proteins
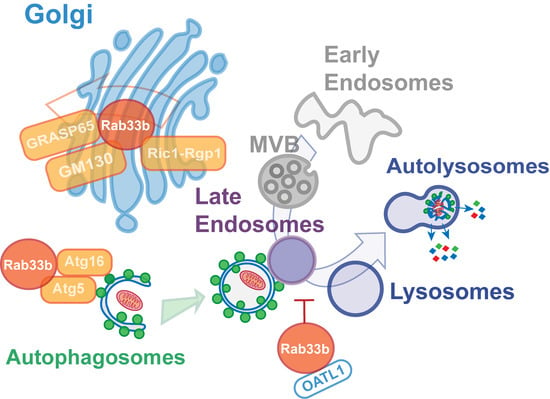
4. Membrane Trafficking Roles of Rab33b
5. Involvement of Rab33b Function in Multiple Steps in the Autophagic Process
6. An Alternative Role for Rab33 in Unconventional Secretion?
7. Disease and Future Perspectives
Funding
Conflicts of Interest
References
- Viotti, C. ER to Golgi-Dependent Protein Secretion: The Conventional Pathway. Methods Mol. Biol. 2016, 1459, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Nickel, W.; Rabouille, C. Mechanisms of Regulated Unconventional Protein Secretion. Nat. Rev. Mol. Cell Biol. 2009, 10, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Ponpuak, M.; Mandell, M.A.; Kimura, T.; Chauhan, S.; Cleyrat, C.; Deretic, V. Secretory Autophagy. Curr. Opin. Cell Biol. 2015, 35, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Rabouille, C. Pathways of Unconventional Protein Secretion. Trends Cell Biol. 2017, 27, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R. Rab GTPases: Master Regulators That Establish the Secretory and Endocytic Pathways. Mol. Biol. Cell 2017, 28, 712–715. [Google Scholar] [CrossRef] [PubMed]
- Stenmark, H. Rab GTPases as Coordinators of Vesicle Traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef]
- Wennerberg, K. The Ras Superfamily at a Glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Leal, J.B.; Seabra, M.C. Evolution of the Rab Family of Small GTP-Binding Proteins. J. Mol. Biol. 2001, 313, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Bonifacino, J.S.; Glick, B.S. The Mechanisms of Vesicle Budding and Fusion. Cell 2004, 116, 153–166. [Google Scholar] [CrossRef] [Green Version]
- Bastin, G.; Heximer, S.P. Rab Family Proteins Regulate the Endosomal Trafficking and Function of RGS4. J. Biol. Chem. 2013, 288, 21836–21849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jovic, M.; Sharma, M.; Rahajeng, J.; Caplan, S. The Early Endosome: A Busy Sorting Station for Proteins at the Crossroads. Histol. Histopathol. 2010, 25, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Ao, X.; Zou, L.; Wu, Y. Regulation of Autophagy by the Rab GTPase Network. Cell Death Differ. 2014, 21, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of Cells and Tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and Diversity in Autophagy Mechanisms: Lessons from Yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.A.; Yoshimori, T.; Tooze, S.A. The Autophagosome: Origins Unknown, Biogenesis Complex. Nat. Rev. Mol. Cell Biol. 2013, 14, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Bento, C.F.; Renna, M.; Ghislat, G.; Puri, C.; Ashkenazi, A.; Vicinanza, M.; Menzies, F.M.; Rubinsztein, D.C. Mammalian Autophagy: How Does It Work? Annu. Rev. Biochem. 2016, 85, 685–713. [Google Scholar] [CrossRef] [PubMed]
- Bento, C.F.; Puri, C.; Moreau, K.; Rubinsztein, D.C. The Role of Membrane-Trafficking Small GTPases in the Regulation of Autophagy. J. Cell Sci. 2013, 126, 1059–1069. [Google Scholar] [CrossRef]
- Russell, R.C.; Yuan, H.X.; Guan, K.L. Autophagy Regulation by Nutrient Signaling. Cell Res. 2014, 24, 42–57. [Google Scholar] [CrossRef]
- Anding, A.L.; Baehrecke, E.H. Cleaning House: Selective Autophagy of Organelles. Dev. Cell 2017, 41, 10–22. [Google Scholar] [CrossRef] [Green Version]
- da Silva Siqueira, M.; de Moraes Ribeiro, R.; Travassos, L.H. Autophagy and Its Interaction with Intracellular Bacterial Pathogens. Front. Immunol. 2018, 9, 935. [Google Scholar] [CrossRef]
- Czarny, P.; Pawlowska, E.; Bialkowska-Warzecha, J.; Kaarniranta, K.; Blasiak, J. Autophagy in DNA Damage Response. Int. J. Mol. Sci. 2015, 16, 2641–2662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashid, H.-O.; Yadav, R.K.; Kim, H.-R.; Chae, H.-J. ER Stress: Autophagy Induction, Inhibition and Selection. Autophagy 2015, 11, 1956–1977. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. MTOR Signaling in Growth Control and Disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Zachari, M.; Ganley, I.G. The Mammalian ULK1 Complex and Autophagy Initiation. Essays Biochem. 2017, 61, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome Formation from Membrane Compartments Enriched in Phosphatidylinositol 3-Phosphate and Dynamically Connected to the Endoplasmic Reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef] [PubMed]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a Mammalian Homologue of Yeast Apg8p, Is Localized in Autophagosome Membranes after Processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef]
- Fujita, N.; Itoh, T.; Omori, H.; Fukuda, M.; Noda, T.; Yoshimori, T. The Atg16L Complex Specifies the Site of LC3 Lipidation for Membrane Biogenesis in Autophagy. Mol. Biol. Cell 2008, 19, 2092–2100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanada, T.; Noda, N.N.; Satomi, Y.; Ichimura, Y.; Fujioka, Y.; Takao, T.; Inagaki, F.; Ohsumi, Y. The Atg12-Atg5 Conjugate Has a Novel E3-like Activity for Protein Lipidation in Autophagy. J. Biol. Chem. 2007, 282, 37298–37302. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Liu, M.; Li, X.; Liu, J.; Li, H. Origin of the Autophagosome Membrane in Mammals. Biomed. Res. Int. 2018, 2018, 1–9. [Google Scholar] [CrossRef]
- Behrends, C.; Sowa, M.E.; Gygi, S.P.; Harper, J.W. Network Organization of the Human Autophagy System. Nature 2010, 466, 68–76. [Google Scholar] [CrossRef]
- Ravikumar, B.; Sarkar, S.; Davies, J.E.; Futter, M.; Garcia-Arencibia, M.; Green-Thompson, Z.W.; Jimenez-Sanchez, M.; Korolchuk, V.I.; Lichtenberg, M.; Luo, S.; et al. Regulation of Mammalian Autophagy in Physiology and Pathophysiology. Physiol. Rev. 2010, 90, 1383–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoppino, F.C.M.; Militello, R.D.; Slavin, I.; Álvarez, C.; Colombo, M.I. Autophagosome Formation Depends on the Small GTPase Rab1 and Functional ER Exit Sites. Traffic 2010, 11, 1246–1261. [Google Scholar] [CrossRef] [PubMed]
- Winslow, A.R.; Chen, C.-W.; Corrochano, S.; Acevedo-Arozena, A.; Gordon, D.E.; Peden, A.A.; Lichtenberg, M.; Menzies, F.M.; Ravikumar, B.; Imarisio, S.; et al. α-Synuclein Impairs Macroautophagy: Implications for Parkinson’s Disease. J. Cell Biol. 2010, 190, 1023–1037. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, Y.; Ohashi, R.; Kawamura, T.; Iwanari, H.; Kodama, T.; Naito, M.; Hamakubo, T. Phosphatidylinositol 3-Phosphatase Myotubularin-Related Protein 6 (MTMR6) Is Regulated by Small GTPase Rab1B in the Early Secretory and Autophagic Pathways. J. Biol. Chem. 2013, 288, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Webster, C.P.; Smith, E.F.; Bauer, C.S.; Moller, A.; Hautbergue, G.M.; Ferraiuolo, L.; Myszczynska, M.A.; Higginbottom, A.; Walsh, M.J.; Whitworth, A.J.; et al. The C9orf72 Protein Interacts with Rab1a and the ULK1 Complex to Regulate Initiation of Autophagy. EMBO J. 2016, 35, 1656–1676. [Google Scholar] [CrossRef] [PubMed]
- Spang, N.; Feldmann, A.; Huesmann, H.; Bekbulat, F.; Schmitt, V.; Hiebel, C.; Koziollek-Drechsler, I.; Clement, A.M.; Moosmann, B.; Jung, J.; et al. RAB3GAP1 and RAB3GAP2 Modulate Basal and Rapamycin-Induced Autophagy. Autophagy 2014, 10, 2297–2309. [Google Scholar] [CrossRef]
- Talaber, G.; Miklossy, G.; Oaks, Z.; Liu, Y.; Tooze, S.A.; Chudakov, D.M.; Banki, K.; Perl, A. HRES-1/Rab4 Promotes the Formation of LC3+ Autophagosomes and the Accumulation of Mitochondria during Autophagy. PLoS ONE 2014, 9, e84392. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Imarisio, S.; Sarkar, S.; O’Kane, C.J.; Rubinsztein, D.C. Rab5 Modulates Aggregation and Toxicity of Mutant Huntingtin through Macroautophagy in Cell and Fly Models of Huntington Disease. J. Cell Sci. 2008, 121, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Bridges, D.; Fisher, K.; Zolov, S.N.; Xiong, T.; Inoki, K.; Weisman, L.S.; Saltiel, A.R. Rab5 Proteins Regulate Activation and Localization of Target of Rapamycin Complex 1. J. Biol. Chem. 2012, 287, 20913–20921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dou, Z.; Pan, J.A.; Dbouk, H.A.; Ballou, L.M.; DeLeon, J.L.; Fan, Y.; Chen, J.S.; Liang, Z.; Li, G.; Backer, J.M.; et al. Class IA PI3K P110β Subunit Promotes Autophagy through Rab5 Small GTPase in Response to Growth Factor Limitation. Mol. Cell 2013, 50, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Nishida, Y.; Arakawa, S.; Fujitani, K.; Yamaguchi, H.; Mizuta, T.; Kanaseki, T.; Komatsu, M.; Otsu, K.; Tsujimoto, Y.; Shimizu, S. Discovery of Atg5/Atg7-Independent Alternative Macroautophagy. Nature 2009, 533, 130. [Google Scholar] [CrossRef] [PubMed]
- Longatti, A.; Lamb, C.A.; Razi, M.; Yoshimura, S.I.; Barr, F.A.; Tooze, S.A. TBC1D14 Regulates Autophagosome Formation via Rab11- and ULK1-Positive Recycling Endosomes. J. Cell Biol. 2012, 197, 659–675. [Google Scholar] [CrossRef] [PubMed]
- Puri, C.; Vicinanza, M.; Ashkenazi, A.; Gratian, M.J.; Zhang, Q.; Bento, C.F.; Renna, M.; Menzies, F.M.; Rubinsztein, D.C. The RAB11A-Positive Compartment Is a Primary Platform for Autophagosome Assembly Mediated by WIPI2 Recognition of PI3P-RAB11A. Dev. Cell 2018, 45, 114–131. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Fukuda, M. Rab12 Regulates MTORC1 Activity and Autophagy through Controlling the Degradation of Amino-Acid Transporter PAT4. EMBO Rep. 2013, 14, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Itoh, T.; Fukuda, M. Small GTPase Rab12 Regulates Constitutive Degradation of Transferrin Receptor. Traffic 2011, 12, 1432–1443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Dai, F.; Cui, L.Q.; Zhou, B.; Guo, Y.Q. Up-Regulation of the Active Form of Small GTPase Rab13 Promotes Macroautophagy in Vascular Endothelial Cells. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Mauvezin, C.; Neisch, A.L.; Ayala, C.I.; Kim, J.; Beltrame, A.; Braden, C.R.; Gardner, M.K.; Hays, T.S.; Neufeld, T.P. Coordination of Autophagosome–lysosome Fusion and Transport by a Klp98A–Rab14 Complex in Drosophila. J. Cell Sci. 2016, 129, 971–982. [Google Scholar] [CrossRef]
- Binotti, B.; Pavlos, N.J.; Riedel, D.; Wenzel, D.; Vorbrüggen, G.; Schalk, A.M.; Kühnel, K.; Boyken, J.; Erck, C.; Martens, H.; et al. The GTPase Rab26 Links Synaptic Vesicles to the Autophagy Pathway. Elife 2015, 4, e05597. [Google Scholar] [CrossRef]
- Hirota, Y.; Tanaka, Y. A Small GTPase, Human Rab32, Is Required for the Formation of Autophagic Vacuoles under Basal Conditions. Cell. Mol. Life Sci. 2009, 66, 2913–2932. [Google Scholar] [CrossRef]
- Itoh, T.; Fujita, N.; Kanno, E.; Yamamoto, A.; Yoshimori, T.; Fukuda, M. Golgi-Resident Small GTPase Rab33B Interacts with Atg16L and Modulates Autophagosome Formation. Mol. Biol. Cell 2008, 19, 2916–2925. [Google Scholar] [CrossRef]
- Seto, S.; Sugaya, K.; Tsujimura, K.; Nagata, T.; Horii, T.; Koide, Y. Rab39a Interacts with Phosphatidylinositol 3-Kinase and Negatively Regulates Autophagy Induced by Lipopolysaccharide Stimulation in Macrophages. PLoS ONE 2013, 8, e83324. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Huang, W.; Lin, T.H.; Groulx, J.F.; Jean, S.; Nguyen, J.; Kuchitsu, Y.; Koyama-Honda, I.; Mizushima, N.; Fukuda, M.; et al. Genetic Screen in Drosophila Muscle Identifies Autophagy-Mediated T-Tubule Remodeling and a Rab2 Role in Autophagy. Elife 2017. [Google Scholar] [CrossRef] [PubMed]
- Jager, S. Role for Rab7 in Maturation of Late Autophagic Vacuoles. J. Cell Sci. 2004, 117, 4837–4848. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Lee, J.S.; Inn, K.S.; Gack, M.U.; Li, Q.; Roberts, E.A.; Vergne, I.; Deretic, V.; Feng, P.; Akazawa, C.; et al. Beclin1-Binding UVRAG Targets the Class C Vps Complex to Coordinate Autophagosome Maturation and Endocytic Trafficking. Nat. Cell Biol. 2008, 10, 776–787. [Google Scholar] [CrossRef]
- Pankiv, S.; Alemu, E.A.; Brech, A.; Bruun, J.A.; Lamark, T.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. FYCO1 Is a Rab7 Effector That Binds to LC3 and PI3P to Mediate Microtubule plus End-Directed Vesicle Transport. J. Cell Biol. 2010, 188, 253–269. [Google Scholar] [CrossRef] [PubMed]
- Tabata, K.; Matsunaga, K.; Sakane, A.; Sasaki, T.; Noda, T.; Yoshimori, T. Rubicon and PLEKHM1 Negatively Regulate the Endocytic/Autophagic Pathway via a Novel Rab7-Binding Domain. Mol. Biol. Cell 2010, 21, 4162–4172. [Google Scholar] [CrossRef]
- Yu, L.; McPhee, C.K.; Zheng, L.; Mardones, G.A.; Rong, Y.; Peng, J.; Mi, N.; Zhao, Y.; Liu, Z.; Wan, F.; et al. Termination of Autophagy and Reformation of Lysosomes Regulated by MTOR. Nature 2010, 465, 942–946. [Google Scholar] [CrossRef]
- Fader, C.M.; Sánchez, D.; Furlán, M.; Colombo, M.I. Induction of Autophagy Promotes Fusion of Multivesicular Bodies with Autophagic Vacuoles in K562 Cells. Traffic 2008, 9, 230–250. [Google Scholar] [CrossRef]
- Szatmari, Z.; Kis, V.; Lippai, M.; Hegedus, K.; Farago, T.; Lorincz, P.; Tanaka, T.; Juhasz, G.; Sass, M. Rab11 Facilitates Cross-Talk between Autophagy and Endosomal Pathway through Regulation of Hook Localization. Mol. Biol. Cell 2014, 25, 522–531. [Google Scholar] [CrossRef]
- Jean, S.; Cox, S.; Nassari, S.; Kiger, A.A. Starvation-Induced MTMR13 and RAB21 Activity Regulates VAMP8 to Promote Autophagosome-Lysosome Fusion. EMBO Rep. 2015, 16, 297–311. [Google Scholar] [CrossRef]
- Munafó, D.B.; Colombo, M.I. Induction of Autophagy Causes Dramatic Changes in the Subcellular Distribution of GFP-Rab24. Traffic 2002, 3, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Tambe, Y.; Yamamoto, A.; Isono, T.; Chano, T.; Fukuda, M.; Inoue, H. The Drs Tumor Suppressor Is Involved in the Maturation Process of Autophagy Induced by Low Serum. Cancer Lett. 2009, 283, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Ylä-Anttila, P.; Mikkonen, E.; Happonen, K.E.; Holland, P.; Ueno, T.; Simonsen, A.; Eskelinen, E.L. RAB24 Facilitates Clearance of Autophagic Compartments during Basal Conditions. Autophagy 2015, 11, 1833–1848. [Google Scholar] [CrossRef] [PubMed]
- Amaya, C.; Militello, R.D.; Calligaris, S.D.; Colombo, M.I. Rab24 Interacts with the Rab7/Rab Interacting Lysosomal Protein Complex to Regulate Endosomal Degradation. Traffic 2016, 17, 1181–1196. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Kanno, E.; Uemura, T.; Waguri, S.; Fukuda, M. OATL1, a Novel Autophagosome-Resident Rab33B-GAP, Regulates Autophagosomal Maturation. J. Cell Biol. 2011, 192, 838–853. [Google Scholar] [CrossRef]
- Fader, C.M.; Colombo, M.I. Autophagy and Multivesicular Bodies: Two Closely Related Partners. Cell Death Differ. 2009, 16, 70–78. [Google Scholar] [CrossRef]
- Ding, X.; Jiang, X.; Tian, R.; Zhao, P.; Li, L.; Wang, X.; Chen, S.; Zhu, Y.; Mei, M.; Bao, S.; et al. RAB2 Regulates the Formation of Autophagosome and Autolysosome in Mammalian Cells. Autophagy 2019, 1–13. [Google Scholar] [CrossRef]
- Zheng, J.Y.; Koda, T.; Fujiwara, T.; Kishi, M.; Ikehara, Y.; Kakinuma, M. A Novel Rab GTPase, Rab33B, Is Ubiquitously Expressed and Localized to the Medial Golgi Cisternae. J. Cell Sci. 1998, 111, 1061–1069. [Google Scholar]
- Zheng, J.Y.; Koda, T.; Arimura, Y.; Kishi, M.; Kakinuma, M. Structure and Expression of the Mouse S10 Gene. Biochim. Et Biophys. Acta Gene Struct. Expr. 1997, 1351, 47–50. [Google Scholar] [CrossRef]
- Cheng, E.; Trombetta, S.E.; Kovacs, D.; Beech, R.D.; Ariyan, S.; Reyes-Mugica, M.; McNiff, J.M.; Narayan, D.; Kluger, H.M.; Picardo, M.; et al. Rab33A: Characterization, Expression, and Suppression by Epigenetic Modification. J. Investig. Dermatol. 2006, 126, 2257–2271. [Google Scholar] [CrossRef] [Green Version]
- Koda, T.; Kakinuma, M. Molecular Cloning of a CDNA Encoding a Novel Small GTP-Binding Protein. FEBS Lett. 1993, 328, 21–24. [Google Scholar] [CrossRef]
- Nakazawa, H.; Sada, T.; Toriyama, M.; Tago, K.; Sugiura, T.; Fukuda, M.; Inagaki, N. Rab33a Mediates Anterograde Vesicular Transport for Membrane Exocytosis and Axon Outgrowth. J. Neurosci. 2012, 32, 12712–12725. [Google Scholar] [CrossRef] [Green Version]
- Zografou, S.; Basagiannis, D.; Papafotika, A.; Shirakawa, R.; Horiuchi, H.; Auerbach, D.; Fukuda, M.; Christoforidis, S. A Complete Rab Screening Reveals Novel Insights in Weibel-Palade Body Exocytosis. J. Cell Sci. 2012, 125, 4780–4790. [Google Scholar] [CrossRef]
- Imai, A.; Tsujimura, M.; Yoshie, S.; Fukuda, M. The Small GTPase Rab33A Participates in Regulation of Amylase Release from Parotid Acinar Cells. Biochem. Biophys. Res. Commun. 2015, 461, 469–474. [Google Scholar] [CrossRef]
- Jiang, S.; Storrie, B. Cisternal Rab Proteins Regulate Golgi Apparatus Redistribution in Response to Hypotonic Stress. Mol. Biol. Cell 2005, 16, 2586–2596. [Google Scholar] [CrossRef] [Green Version]
- Valsdottir, R.; Hashimoto, H.; Ashman, K.; Koda, T.; Storrie, B.; Nilsson, T. Identification of Rabaptin-5, Rabex-5, and GM130 as Putative Effectors of Rab33b, a Regulator of Retrograde Traffic between the Golgi Apparatus and ER. FEBS Lett. 2001, 508, 201–209. [Google Scholar] [CrossRef]
- Pelham, H.R. Recycling of Proteins between the Endoplasmic Reticulum and Golgi Complex. Curr. Opin. Cell Biol. 1991, 3, 585–591. [Google Scholar] [CrossRef]
- Heffernan, L.F.; Simpson, J.C. The Trials and Tubule-Ations of Rab6 Involvement in Golgi-to-ER Retrograde Transport. Biochem. Soc. Trans. 2014, 42, 1453–1459. [Google Scholar] [CrossRef]
- Starr, T.; Sun, Y.; Wilkins, N.; Storrie, B. Rab33b and Rab6 Are Functionally Overlapping Regulators of Golgi Homeostasis and Trafficking. Traffic 2010, 11, 626–636. [Google Scholar] [CrossRef]
- Liu, S.; Storrie, B. Are Rab Proteins the Link between Golgi Organization and Membrane Trafficking? Cell. Mol. Life Sci. 2012, 69, 4093–4106. [Google Scholar] [CrossRef]
- Pusapati, G.V.; Luchetti, G.; Pfeffer, S.R. Ric1-Rgp1 Complex Is a Guanine Nucleotide Exchange Factor for the Late Golgi Rab6A GTPase and an Effector of the Medial Golgi Rab33B GTPase. J. Biol. Chem. 2012, 287, 42129–42137. [Google Scholar] [CrossRef] [Green Version]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [Green Version]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the Human Interactome Defines Protein Communities and Disease Networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef]
- Geppert, M.; Südhof, T.C. RAB3 AND SYNAPTOTAGMIN: The Yin and Yang of Synaptic Membrane Fusion. Annu. Rev. Neurosci. 2002, 21, 75–95. [Google Scholar] [CrossRef]
- Vadlamudi, R.K.; Wang, R.-A.; Talukder, A.H.; Adam, L.; Johnson, R.; Kumar, R. Evidence of Rab3A Expression, Regulation of Vesicle Trafficking, and Cellular Secretion in Response to Heregulin in Mammary Epithelial Cells. Mol. Cell. Biol. 2002, 20, 9092–9101. [Google Scholar] [CrossRef]
- Bustos, M.A.; Lucchesi, O.; Ruete, M.C.; Mayorga, L.S.; Tomes, C.N. Rab27 and Rab3 Sequentially Regulate Human Sperm Dense-Core Granule Exocytosis. Proc. Natl. Acad. Sci. USA 2012, 109, E2057–E2066. [Google Scholar] [CrossRef]
- Plutner, H.; Cox, A.D.; Pind, S.; Khosravi-Far, R.; Bourne, J.R.; Schwaninger, R.; Der, C.J.; Balch, W.E. Rab1b Regulates Vesicular Transport between the Endoplasmic Reticulum and Successive Golgi Compartments. J. Cell Biol. 1991, 115, 31–43. [Google Scholar] [CrossRef]
- Kakuta, S.; Yamaguchi, J.; Suzuki, C.; Sasaki, M.; Kazuno, S.; Uchiyama, Y. Small GTPase Rab1B Is Associated with ATG9A Vesicles and Regulates Autophagosome Formation. FASEB J. 2017, 31, 3757–3773. [Google Scholar] [CrossRef]
- Bexiga, M.G.; Simpson, J.C. Human Diseases Associated with Form and Function of the Golgi Complex. Int. J. Mol. Sci. 2013, 14, 18670–18681. [Google Scholar] [CrossRef]
- Mizushima, N.; Yamamoto, A.; Hatano, M.; Kobayashi, Y.; Kabey, Y.; Suzuki, K.; Tokuhis, T.; Ohsumi, Y.; Yoshimori, T. Dissection of Autophagosome Formation Using Apg5-Deficient Mouse Embryonic Stem Cells. J. Cell Biol. 2001, 152, 657–667. [Google Scholar] [CrossRef]
- Mizushima, N.; Sugita, H.; Yoshimori, T.; Ohsumi, Y. A New Protein Conjugation System in Human: The Counterpart of the Yeast Apg12p Conjugation System Essential for Autophagy. J. Biol. Chem. 1998, 273, 33889–33892. [Google Scholar] [CrossRef]
- Mizushima, N.; Kuma, A.; Kobayashi, Y.; Yamamoto, A.; Matsubae, M.; Takao, T.; Natsume, T.; Ohsumi, Y.; Yoshimori, T. Mouse Apg16L, a Novel WD-Repeat Protein, Targets to the Autophagic Isolation Membrane with the Apg12-Apg5 Conjugate. J. Cell Sci. 2003, 116, 1679–1688. [Google Scholar] [CrossRef]
- Panarella, A.; Bexiga, M.G.; Galea, G.; O’ Neill, E.D.; Salvati, A.; Dawson, K.A.; Simpson, J.C. A Systematic High-Content Screening Microscopy Approach Reveals Key Roles for Rab33b, OATL1 and Myo6 in Nanoparticle Trafficking in HeLa Cells. Sci. Rep. 2016, 6, 28865. [Google Scholar] [CrossRef]
- Abrahamsen, H. Stenmark, H. Protein Secretion: Unconventional Exit by Exophagy. Curr. Biol. 2010, 20, PR415–R418. [Google Scholar] [CrossRef]
- Ishibashi, K.; Uemura, T.; Waguri, S.; Fukuda, M. Atg16L1, an Essential Factor for Canonical Autophagy, Participates in Hormone Secretion from PC12 Cells Independently of Autophagic Activity. Mol. Biol. Cell 2012, 23, 3193–3202. [Google Scholar] [CrossRef]
- Döring, T.; Prange, R. Rab33B and Its Autophagic Atg5/12/16L1 Effector Assist in Hepatitis B Virus Naked Capsid Formation and Release. Cell. Microbiol. 2015, 17, 747–764. [Google Scholar] [CrossRef]
- Odorizzi, G. The Multiple Personalities of Alix. J. Cell Sci. 2006, 119, 3025–3032. [Google Scholar] [CrossRef]
- Alshammari, M.J.; Al-Otaibi, L.; Alkuraya, F.S. Mutation in RAB33B, Which Encodes a Regulator of Retrograde Golgi Transport, Defines a Second Dyggve-Melchior-Clausen Locus. J. Med. Genet. 2012, 49, 455–461. [Google Scholar] [CrossRef]
- Dupuis, N.; Lebon, S.; Kumar, M.; Drunat, S.; Graul-Neumann, L.M.; Gressens, P.; El Ghouzzi, V. A Novel RAB33B Mutation in Smith-McCort Dysplasia. Hum. Mutat. 2013, 34, 283–286. [Google Scholar] [CrossRef]
- Salian, S.; Cho, T.-J.; Phadke, S.R.; Gowrishankar, K.; Bhavani, G.S.; Shukla, A.; Jagadeesh, S.; Kim, O.-H.; Nishimura, G.; Girisha, K.M. Additional Three Patients with Smith-McCort Dysplasia Due to Novel RAB33B Mutations. Am. J. Med. Genet. Part A 2017, 17, 588–595. [Google Scholar] [CrossRef]
- Nakamura, K.; Kurokawa, T.; Nagano, A.; Nakamura, S.; Taniguchi, K.; Hamazaki, M. Dyggve-Melchior-Clausen Syndrome without Mental Retardation (Smith-McCort Dysplasia): Morphological Findings in the Growth Plate of the Iliac Crest. Am. J. Med. Genet. 1997, 72, 11–17. [Google Scholar] [CrossRef]
- Huang, J.; Klionsky, D.J. Autophagy and Human Disease. Cell Cycle 2007, 24, 1837–1849. [Google Scholar] [CrossRef]
- Zappa, F.; Failli, M.; De Matteis, M.A. The Golgi Complex in Disease and Therapy. Curr. Opin. Cell Biol. 2018, 50, 102–116. [Google Scholar] [CrossRef]
- Kim, J.; Gee, H.Y.; Lee, M.G. Unconventional Protein Secretion–New Insights into the Pathogenesis and Therapeutic Targets of Human Diseases. J. Cell Sci. 2018, 131, jcs213686. [Google Scholar] [CrossRef]


| Mammalian Rab GTPases That Function in the Autophagic Pathway Contributing to Autophagosome Biogenesis | |||
| Rab Protein | Autophagy Effectors/Interactors | Function in Autophagy | References |
| Rab1a Rab1b | Atg5, ULK1 Unknown | Translocation of ULK1 complex and mAtg9-containing vesicles at pre-autophagosomal membranes is Rab1a-dependent. Activity of Rab1b is required for autophagosome formation at ER exit sites, it may regulate the amount of PI(3)P in the omegasome through interaction with myotubularin-related protein 6. | [30,31,32,33,34,35] |
| Rab3b Rab3d | LC3 Atg16L | The GTPase-activating domain of RAB3GAP1/2 cooperates with Atg3 or Atg16L to sustain autophagosome biogenesis. Indirect evidence for involvement of Rab3 in autophagosome biogenesis. | [30,36] |
| Rab4a | Unknown | Formation of LC3-positive autophagic structures in response to overexpression of Rab4, following localisation to those structures upon blockade of mTORC1. | [37] |
| Rab5c Rab5 | LC3, Atg10, PIK3C3 | Rab5 acts as an activator of the Vps34/Beclin1-PIK3C3 complex and promotes Atg5-Atg12 conjugation, which in turn leads to elongation of pre-autophagosomal structures. Rab5 forms part of a signalling cascade that promotes initiation of autophagy independently of nutrient shortage and controls mTORC1 activation and localisation. | [30,38,39,40] |
| Rab9a | Unknown | Rab9a function is required for generation of autophagosomes from trans-Golgi-derived IMs in Atg5- and Atg7-independent autophagy. | [41] |
| Rab11 | MLST8, TBC1D14, ULK1, Atg16L | Rab11 mediates incorporation of recycling endosomal membranes that contain ULK1 and mAtg9 to the IM and modulates autophagosome elongation upon amino acid starvation. This process is negatively regulated by the non-GAP effector TBC1D14. Rab11a-positive membranes provide a platform for autophagosome biogenesis by favouring the recruitment of the Atg16L complex. | [30,42,43] |
| Rab12 | Unknown | Regulates trafficking and lysosomal degradation of the amino-acid transporter PAT4. Loss of Rab12 results in accumulation of PAT4 and increased mTORC1 activity, which thereby inhibits autophagy. | [44,45] |
| Rab13 | Unknown | Mediates pterostilbene-induced autophagy in endothelial cells via functional interaction of GTP-active form with growth factor receptor-bound protein 2 (Grb2), which leads to mTOR inhibition. | [46] |
| Rab14 | Unknown | Functions in earlier stages of autophagosome formation; its silencing causes a reduction in the size of autophagic vesicles, whereas overexpression leads to the opposite effect. | [47] |
| Rab26 | Atg16L | The GTP-form of Rab26 selectively recruits Atg16L and Rab33b into large clusters of synaptic vesicles that represent pre-autophagosomal compartments. | [48] |
| Rab32 | Unknown | Rab32 facilitates the formation of LC3-positive autophagic structures from the ER membrane during basal autophagy. | [49] |
| Rab33b | Atg5, Atg16L | Regulates conjugation of LC3 to PE through recruitment of the Atg12-Atg5-Atg16L complex. | [30,50] |
| Rab39a | Atg5, Atg14L, PIK3C3, Beclin, Vps34 | Negatively regulates autophagy induced by LPS in macrophages through PI3K/Beclin-dependent mechanisms. | [30,51] |
| Mammalian Rab GTPases That Function in the Autophagic Pathway Contributing to Autophagosome Maturation | |||
| RAB Protein | Autophagy Effectors/Interactors | Function in Autophagy | References |
| Rab2a/b | HOPS complex | Promotes autophagosome clearance via its localisation to autophagosomes. Mediates trans-SNARE complex formation and coordinates fusion of amphisomes through the HOPS complex with Rab7-marked structures. | [52] |
| Rab7 | UVRAG, RILP, FYCO1, CLN3, Rubicon, PIK3C2A, UBE1DC1 | Main regulator of trafficking of autophagosomes and their fusion to lysosomes via effector proteins; binding to LC3 and PI(3)P through FYCO1 regulates Rab7-dependent transport of autophagosomes through microtubule tracks; RILP mediates binding to dynactin-dynein1; the component of the Beclin 1 complex, UVRAG, activates Rab7 through the GEF activity of HOPS complex; Rubicon inhibits Rab7 activation by blocking UVRAG function. | [30,53,54,55,56,57] |
| Rab11 | Hook | Regulation at the level of fusion between autophagosomes and multivesicular bodies. Drosophila Rab11 removes the microtubule binding protein Hook, a negative regulator of endosome maturation, allowing subsequent fusion events. | [58,59] |
| Rab14 | Klp98A | Through its effector Klp98A (Drosophila orthologue of human KIF16B kinesin 3 family member), Rab14 controls the positioning of lysosomes and promotes autophagosome-lysosome function. | [47] |
| Rab21 | UBE1DC1, VAMP7, VAMP8 | Rab21 endosomal activity promotes VAMP8 endo-lysosomal trafficking to Rab7-positive late endosomes and SNARE-mediated autophagosome-lysosome fusion, which results enhanced in response to starvation. | [30,60] |
| Rab24 | Drs, Rab7, RILP | Following induction of autophagy Rab24 localises in spots decorated with LC3, mediating the clearance of late autophagic compartments after their acquisition of degradative capacity and upon nutrient-rich conditions. Its interaction with drs tumour suppressor regulates fusion with lysosomes. Interacts with Rab7/RILP | [61,62,63,64] |
| Rab33b | UVRAG, CLN3 | Regulates the fusion of autophagosomes with lysosomes. Regulation by its GAP protein (OATL1) is necessary to ensure autophagosome maturation. | [30,65] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morgan, N.E.; Cutrona, M.B.; Simpson, J.C. Multitasking Rab Proteins in Autophagy and Membrane Trafficking: A Focus on Rab33b. Int. J. Mol. Sci. 2019, 20, 3916. https://doi.org/10.3390/ijms20163916
Morgan NE, Cutrona MB, Simpson JC. Multitasking Rab Proteins in Autophagy and Membrane Trafficking: A Focus on Rab33b. International Journal of Molecular Sciences. 2019; 20(16):3916. https://doi.org/10.3390/ijms20163916
Chicago/Turabian StyleMorgan, Niamh E., Meritxell B. Cutrona, and Jeremy C. Simpson. 2019. "Multitasking Rab Proteins in Autophagy and Membrane Trafficking: A Focus on Rab33b" International Journal of Molecular Sciences 20, no. 16: 3916. https://doi.org/10.3390/ijms20163916
APA StyleMorgan, N. E., Cutrona, M. B., & Simpson, J. C. (2019). Multitasking Rab Proteins in Autophagy and Membrane Trafficking: A Focus on Rab33b. International Journal of Molecular Sciences, 20(16), 3916. https://doi.org/10.3390/ijms20163916