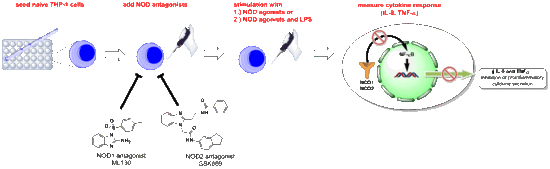
THP-1 Cells and Pro-Inflammatory Cytokine Production: An In Vitro Tool for Functional Characterization of NOD1/NOD2 Antagonists
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect of Selected Chemicals on Cell Viability
2.2. Determination of Optimal Experimental Conditions for Screening (Dose-Finding Study)
2.3. Effect of NOD Antagonists on IL-8 Secretion from Stimulated THP-1 Cells
2.4. Effect of Selected NOD1/NOD2 Antagonists on NOD1/2-TLR4 Synergy
3. Materials and Methods
3.1. Test Chemicals
3.2. Cell Culture
3.3. Cell Viability
3.4. Cytokine Production (ELISA)
3.5. Data Analysis and Statistics
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| C12-iE-DAP | lauroyl-γ-d-glutamyl-meso-diaminopimelic acid |
| h-CLAT | human cell line activation test |
| iE-DAP | γ-d-glutamyl-meso-diaminopimelic acid |
| IL | interleukin |
| LDH | lactate dehydrogenase |
| LPS | lipopolysaccharide |
| MDP | muramyl dipeptide |
| MTS | 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium |
| NF-κB | nuclear factor κB |
| NLR | NOD-like receptor |
| NOD | nucleotide-binding oligomerization domain |
| PRR | pattern recognition receptors |
| SEAP | secreted embryonic alkaline phosphatase |
| TLR | toll-like receptor |
| TNF-α | tumor necrosis factor-α |
Appendix A


References
- Fritz, J.H.; Ferrero, R.L.; Philpott, D.J.; Girardin, S.E. Nod-like Proteins in Immunity, Inflammation and Disease. Nat. Immunol. 2006, 7, 1250–1257. [Google Scholar] [CrossRef] [PubMed]
- Philpott, D.J.; Sorbara, M.T.; Robertson, S.J.; Croitoru, K.; Girardin, S.E. NOD Proteins: Regulators of Inflammation in Health and Disease. Nat. Rev. Immunol. 2014, 14, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Inohara, N.; Nuñez, G. NODs: Intracellular Proteins Involved in Inflammation and Apoptosis. Nat. Rev. Immunol. 2003, 3, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Girardin, S.E.; Boneca, I.G.; Carneiro, L.A.M.; Antignac, A.; Jéhanno, M.; Viala, J.; Tedin, K.; Taha, M.K.; Labigne, A.; ähringer, U.; et al. Nod1 Detects a Unique Muropeptide from Gram-negative Bacterial Peptidoglycan. Science 2003, 300, 1584–1587. [Google Scholar] [CrossRef] [PubMed]
- Girardin, S.E.; Jéhanno, M.; Mengin-Lecreulx, D.; Sansonetti, P.J.; Alzari, P.M.; Philpott, D.J. Identification of the Critical Residues Involved in Peptidoglycan Detection by Nod1. J. Biol. Chem. 2005, 280, 38648–38656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamaillard, M.; Hashimoto, M.; Horie, Y.; Masumoto, J.; Qiu, S.; Saab, L.; Ogura, Y.; Kawasaki, A.; Fukase, K.; Kusumoto, S.; et al. An Essential Role for NOD1 in Host Recognition of Bacterial Peptidoglycan Containing Diaminopimelic acid. Nat. Immunol. 2003, 4, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Inohara, N.; Ogura, Y.; Fontalba, A.; Gutierrez, O.; Pons, F.; Crespo, J.; Fukase, K.; Inamura, S.; Kusumoto, S.; Hashimoto, M.; et al. Host Recognition of Bacterial Muramyl Dipeptide Mediated Through NOD2. J. Biol. Chem. 2003, 278, 5509–5512. [Google Scholar] [CrossRef] [PubMed]
- Girardin, S.E.; Boneca, I.G.; Viala, J.; Chamaillard, M.; Labigne, A.; Thomas, G.; Philpott, D.J.; Sansonetti, P.J. Nod2 is a General Sensor of Peptidoglycan Through Muramyl Dipeptide (MDP) Detection. J. Biol. Chem. 2003, 278, 8869–8872. [Google Scholar] [CrossRef] [PubMed]
- Fritz, J.H.; Girardin, S.E.; Fitting, C.; Werts, C.; Mengin-Lecreulx, D.; Caroff, M.; Cavaillon, J.M.; Philpott, D.J.; Adib-Conguy, M. Synergistic Stimulation of Human Monocytes and Dendritic Cells by Toll-like Receptor 4 and Nod1- and Nod2-Activating Agonists. Eur. J. Immunol. 2005, 35, 2459–2470. [Google Scholar] [CrossRef] [PubMed]
- Uehori, J.; Fukase, K.; Akazawa, T.; Uematsu, S.; Akira, S.; Funami, K.; Shingai, M.; Matsumoto, M.; Azuma, I.; Toyoshima, K.; et al. Dendritic Cell Maturation Induced by Muramy Dipeptide (MDP) Derivatives: Monoacylated MDP Confers TLR2/TLR4 Activation. J. Immunol. 2005, 174, 7096–7103. [Google Scholar] [CrossRef]
- Tada, H.; Aiba, S.; Shibata, K.; Ohteki, T.; Takada, H. Synergistic Effect of Nod1 and Nod2 Agonists with Toll-like receptor Agonists on Human Dendritic cells to Generate Interleukin-12 and T Helper Type 1 Cells. Infect. Immun. 2005, 73, 7967–7976. [Google Scholar] [CrossRef] [PubMed]
- Van Heel, D.A.; Ghosh, S.; Butler, M.; Hunt, K.; Foxwell, B.M.; Mengin-Lecreulx, D.; Playford, R.J. Synergistic Enhancement of Toll-like Receptor Responses by NOD1 Activation. Eur. J. Immunol. 2005, 35, 2471–2476. [Google Scholar] [CrossRef] [PubMed]
- Jakopin, Ž. Nucleotide-Binding Oligomerization Domain (NOD) Inhibitors: A Rational Approach Toward Inhibition of NOD Signaling Pathway. J. Med. Chem. 2014, 57, 6897–6918. [Google Scholar] [CrossRef] [PubMed]
- Correa, R.G.; Milutinovic, S.; Reed, J.C. Roles of NOD1 (NLRC1) and NOD2 (NLRC2) in Innate Immunity and Inflammatory Diseases. Biosci. Rep. 2012, 608, 597–608. [Google Scholar] [CrossRef]
- Moreno, L.; Gatheral, T. Therapeutic Targeting of NOD1 Receptors. Br. J. Pharmacol. 2013, 170, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, G.; Khan, P.; Yuan, H.; Brown, B.; Divlianska, D.B.; Stonich, D.; Peddibhotla, S.; Su, Y.; Dad, S.; Sergienko, E.; et al. High Throughput Screening Assays for NOD1 Inhibitors-Probe 1. Available online: https://www.ncbi.nlm.nih.gov/books/NBK50683/ (accessed on 20 October 2010).
- Magnuson, G.; Khan, P.; Yuan, H.; Brown, B.; Divlianska, D.B.; Stonich, D.; Peddibhotla, S.; Su, Y.; Dad, S.; Sergienko, E.; et al. High Throughput Screening Assays for NOD1 Inhibitors-Probe 2. Available online: https://www.ncbi.nlm.nih.gov/pubmed/21433392 (accessed on 4 October 2010).
- Rickard, D.J.; Sehon, C.A.; Kasparcova, V.; Kallal, L.A.; Haile, P.A.; Zeng, X.; Montoute, M.N.; Poore, D.D.; Li, H.; Wu, Z.; et al. Identification of Selective Small Molecule Inhibitors of the Nucleotide-Binding Oligomerization Domain 1 (NOD1) Signaling Pathway. PLoS ONE 2014, 9, e96737. [Google Scholar] [CrossRef]
- Khan, P.M.; Correa, R.G.; Divlianska, D.B.; Peddibhotla, S.; Sessions, E.H.; Magnuson, G.; Brown, B.; Suyama, E.; Yuan, H.; Mangravita-Novo, A.; et al. Identification of Inhibitors of NOD1-Induced Nuclear Factor-κB Activation. ACS Med. Chem. Lett. 2011, 2, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Correa, R.G.; Khan, P.M.; Askari, N.; Zhai, D.; Gerlic, M.; Brown, B.; Magnuson, G.; Spreafico, R.; Albani, S.; Sergienko, E.; et al. Discovery and Characterization of 2-Aminobenzimidazole Derivatives as Selective NOD1 Inhibitors. Chem. Biol. 2011, 18, 825–832. [Google Scholar] [CrossRef] [Green Version]
- Rickard, D.J.; Sehon, C.A.; Kasparcova, V.; Kallal, L.A.; Zeng, X.; Montoute, M.N.; Chordia, T.; Poore, D.D.; Li, H.; Wu, Z.; et al. Identification of Benzimidazole Diamides as Selective Inhibitors of the Nucleotide-Binding Oligomerization Domain 2 (NOD2) Signaling Pathway. PLoS ONE 2013, 8, e69619. [Google Scholar] [CrossRef]
- Keček Plešec, K.; Urbančič, D.; Gobec, M.; Pekošak, A.; Tomašič, T.; Anderluh, M.; Mlinarič-Raščan, I.; Jakopin, Ž. Identification of Indole Scaffold-Based Dual Inhibitors of NOD1 and NOD2. Bioorg. Med. Chem. 2016, 24, 5221–5234. [Google Scholar] [CrossRef]
- Wang, S.; Yang, J.; Li, X.; Liu, Z.; Wu, Y.; Si, G.; Tao, Y.; Zhao, N.; Hu, X.; Ma, Y.; et al. Discovery of 1,4-Benzodiazepine-2,5-dione (BZD) Derivatives as Dual Nucleotide Binding Oligomerization Domain Containing 1/2 (NOD1/NOD2) Antagonists Sensitizing Paclitaxel (PTX) to Suppress Lewis Lung Carcinoma (LLC) Growth in vivo. J. Med. Chem. 2017, 60, 5162–5192. [Google Scholar] [CrossRef] [PubMed]
- Zurek, B.; Bielig, H.; Kufer, T.A. Cell-Based Reporter Assay to Analyze Activation of Nod1 and Nod2. Methods Mol. Biol. 2011, 748, 107–119. [Google Scholar]
- Zhao, L.; Kwon, M.-J.; Huang, S.; Lee, J.Y.; Fukase, K.; Inohara, N.; Hwang, D.H. Differential Modulation of Nods Signaling Pathways by Fatty Acids in Human Colonic Epithelial HCT116 Cells. J. Biol. Chem. 2007, 282, 11618–11628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva Correia, J.; Miranda, Y.; Austin-Brown, N.; Hsu, J.; Mathison, J.; Xiang, R.; Zhou, H.; Li, Q.; Han, J.; Ulevitch, R.J. Nod1-Dependent Control of Tumor Growth. Proc. Natl. Acad. Sci. USA 2006, 103, 1840–1845. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.C.; Correa, R.G.; Roth, G.P.; Khan, P.M. Modulators of Nod1 and Nod2 signaling, methods of identifying modulators of Nod1 and Nod2 signaling, and uses thereof. U.S. Patent Application No. WO2012/021647A2, 12 February 2012. [Google Scholar]
- Jakopin, Ž.; Corsini, E.; Gobec, M.; Mlinarič-Raščan, I.; Sollner Dolenc, M. Design, Synthesis and Biological Evaluation of Novel Desmuramyldipeptide Analogs. Eur. J. Med. Chem. 2011, 46, 3762–3777. [Google Scholar] [CrossRef]
- Jakopin, Ž.; Gobec, M.; Mlinarič-Raščan, I.; Sollner Dolenc, M. Immunomodulatory Properties of Novel Nucleotide Oligomerization Domain 2 (Nod2) Agonistic Desmuramyldipeptides. J. Med. Chem. 2012, 55, 6478–6488. [Google Scholar] [CrossRef]
- Gobec, M.; Mlinarič-Raščan, I.; Sollner Dolenc, M.; Jakopin, Ž. Structural Requirements of Acylated Gly-l-Ala-d-Glu Analogs for Activation of the Innate Immune Receptor NOD2. Eur. J. Med. Chem. 2016, 116, 1–12. [Google Scholar] [CrossRef]
- Gobec, M.; Tomašič, T.; Štimac, A.; Frkanec, R.; Trontelj, J.; Anderluh, M.; Mlinarič-Raščan, I.; Jakopin, Ž. Discovery of Nanomolar Desmuramylpeptide Agonists of the Innate Immune Receptor Nucleotide-Binding Oligomerization Domain-Containing Protein 2 (NOD2) Possessing Immunostimulatory Properties. J. Med. Chem. 2018, 61, 2707–2724. [Google Scholar] [CrossRef]
- Jakopin, Ž.; Gobec, M.; Kodela, J.; Hazdovac, T.; Mlinarič-Raščan, I.; Sollner Dolenc, M. Synthesis of Conformationally Constrained γ-D-glutamyl-meso-diaminopimelic Acid Derivatives as Ligands of Nucleotide-Binding Oligomerization Domain Protein 1 (Nod1). Eur. J. Med. Chem. 2013, 69, 232–243. [Google Scholar] [CrossRef]
- Mitjans, M.; Galbiati, V.; Lucchi, L.; Viviani, B.; Marinovich, M.; Galli, C.L.; Corsini, E. Use of IL-8 Release and p38 MAPK Activation in THP-1 Cells to Identify Allergens and to Assess Their Potency in Vitro. Toxicol. In Vitro 2010, 24, 1803–1809. [Google Scholar] [CrossRef]
- Mitjans, M.; Viviani, B.; Lucchi, L.; Galli, C.L.; Marinovich, M.; Corsini, E. Role of p38 MAPK in the Selective Release of IL-8 Induced by Chemical Allergen in Naive THP-1 Cells. Toxicol. In Vitro 2008, 22, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 Cell Line: an In Vitro Cell Model for Immune Modulation Approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Uehara, A.; Yang, S.; Fujimoto, Y.; Fukase, K.; Kusumoto, S.; Shibata, K.; Sugawara, S.; Takada, H. Muramyldipeptide and Diaminopimelic Acid-Containing Desmuramylpeptides in Combination with Chemically Synthesized Toll-like Receptor Agonists Synergistically Induced Production of Interleukin-8 in a NOD2- and NOD1-Dependent Manner, Respectively, in Human Monocytic Cells in Culture. Cell Microbiol. 2005, 7, 53–61. [Google Scholar] [PubMed]
- Aldo, P.B.; Craveiro, V.; Guller, S.; Mor, G. Effect of Culture Conditions on the Phenotype of THP-1 Monocyte Cell Line. Am. J. Reprod. Immunol. 2013, 70, 80–86. [Google Scholar] [CrossRef] [Green Version]
- Ashikaga, T.; Sakaguchi, H.; Sono, S.; Kosaka, N.; Ishikawa, M.; Nukada, Y.; Miyazawa, M.; Ito, Y.; Nishiyama, N.; Itagaki, H. A Comparative Evaluation of In Vitro Skin Sensitisation Tests: the Human Cell-Line Activation Test (h-CLAT) Versus the Local Lymph Node Assay (LLNA). Altern. Lab. Anim. 2010, 38, 275–284. [Google Scholar] [CrossRef]
- Tukhvatulin, A.I.; Gitlin, I.I.; Shcheblyakov, D.V.; Artemicheva, N.M.; Burdelya, L.G.; Shmarov, M.M.; Naroditsky, B.S.; Gudkov, A.V.; Gintsburg, A.L.; Logunov, D.Y. Combined Stimulation of Toll-like Receptor 5 and NOD1 Strongly Potentiates Activity of NF-κB, Resulting in Enhanced Innate Immune Reactions and Resistance to Salmonella Enterica Serovar Typhimurium Infection. Infect Immun. 2013, 81, 3855–3864. [Google Scholar] [CrossRef] [PubMed]
- Tukhvatulin, A.I.; Dzharullaeva, A.S.; Tukhvatulina, N.M.; Shcheblyakov, D.V.; Shmarov, M.M.; Dolzhikova, I.V.; Stanhope-Baker, P.; Naroditsky, B.S.; Gudkov, A.V.; Logunov, D.Y.; et al. Powerful Complex Immunoadjuvant Based on Synergistic Effect of Combined TLR4 and NOD2 Activation Significantly Enhances Magnitude of Humoral and Cellular Adaptive Immune Responses. PLoS ONE 2016, 11, e0155650. [Google Scholar] [CrossRef]
- Decker, T.; Lohmann-Matthes, M. A Quick and Simple Method for the Quantitation of Lactate Dehydrogenase Release in Measurements of Cellular Cytotoxicity and Tumor Necrosis Factor (TNF) Activity. J. Immunol. Meth. 1988, 15, 61–69. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jakopin, Ž.; Corsini, E. THP-1 Cells and Pro-Inflammatory Cytokine Production: An In Vitro Tool for Functional Characterization of NOD1/NOD2 Antagonists. Int. J. Mol. Sci. 2019, 20, 4265. https://doi.org/10.3390/ijms20174265
Jakopin Ž, Corsini E. THP-1 Cells and Pro-Inflammatory Cytokine Production: An In Vitro Tool for Functional Characterization of NOD1/NOD2 Antagonists. International Journal of Molecular Sciences. 2019; 20(17):4265. https://doi.org/10.3390/ijms20174265
Chicago/Turabian StyleJakopin, Žiga, and Emanuela Corsini. 2019. "THP-1 Cells and Pro-Inflammatory Cytokine Production: An In Vitro Tool for Functional Characterization of NOD1/NOD2 Antagonists" International Journal of Molecular Sciences 20, no. 17: 4265. https://doi.org/10.3390/ijms20174265
APA StyleJakopin, Ž., & Corsini, E. (2019). THP-1 Cells and Pro-Inflammatory Cytokine Production: An In Vitro Tool for Functional Characterization of NOD1/NOD2 Antagonists. International Journal of Molecular Sciences, 20(17), 4265. https://doi.org/10.3390/ijms20174265