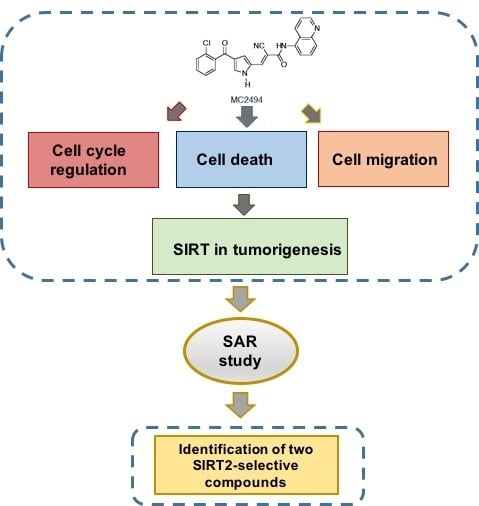
Enzymatic and Biological Characterization of Novel Sirtuin Modulators against Cancer
Abstract
:1. Introduction
2. Results
2.1. MC2494-Related Derivatives and Their Enzymatic Characterization
2.2. Effects of MC2494 Derivatives on Cell Cycle Progression
2.3. Analysis of MC2494 Analogs on Cell Death Regulation in Cancer
2.4. MC2494 Displays Context-Specific Regulation in Cancer Cell Cycle Progression
2.5. MC2494 Regulates Cancer Cell Cycle Progression at Molecular Level
2.6. MC2494 Impacts on Cancer Cell Migration
3. Discussion
4. Materials and Methods
4.1. Ligands
4.2. Morphological Analysis
4.3. Antibodies
4.4. Cell Lines
4.5. SIRT1 Purification
4.6. SIRT Assays
4.7. Cell Cycle and Cell Death Analysis
4.8. Annexin V Staining
4.9. Protein Extraction
4.10. Western Blot
4.11. In Vivo Experiments
4.12. Immunohistochemistry
4.13. Wound-Scratch Assay
4.14. Boyden’s Chamber Assay
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Nebbioso, A.; Tambaro, F.P.; Dell’Aversana, C.; Altucci, L. Cancer epigenetics: Moving forward. PLoS Genet. 2018, 14, e1007362. [Google Scholar] [CrossRef] [PubMed]
- Marks, P.; Rifkind, R.A.; Richon, V.M.; Breslow, R.; Miller, T.; Kelly, W.K. Histone deacetylases and cancer: Causes and therapies. Nat. Rev. Cancer 2001, 1, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Minucci, S.; Pelicci, P.G. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer 2006, 6, 38–51. [Google Scholar] [CrossRef] [PubMed]
- West, A.C.; Johnstone, R.W. New and emerging HDAC inhibitors for cancer treatment. J. Clin. Investig. 2014, 124, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci 2017, 18, 1414. [Google Scholar] [CrossRef] [PubMed]
- Mai, A.; Altucci, L. Epi-drugs to fight cancer: From chemistry to cancer treatment, the road ahead. Int. J. Biochem. Cell Biol. 2009, 41, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Bishayee, A.; Pandey, A.K. Targeting Histone Deacetylases with Natural and Synthetic Agents: An Emerging Anticancer Strategy. Nutrients 2018, 10, 731. [Google Scholar] [CrossRef] [PubMed]
- Haigis, M.C.; Sinclair, D.A. Mammalian sirtuins: Biological insights and disease relevance. Annu. Rev. Pathol. 2010, 5, 253–295. [Google Scholar] [CrossRef] [PubMed]
- Carafa, V.; Nebbioso, A.; Altucci, L. Sirtuins and disease: The road ahead. Front. Pharm. 2012, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Vaquero, A. The conserved role of sirtuins in chromatin regulation. Int. J. Dev. Biol 2009, 53, 303–322. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Redondo, P.; Vaquero, A. The diversity of histone versus nonhistone sirtuin substrates. Genes Cancer 2013, 4, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Carafa, V.; Altucci, L.; Nebbioso, A. Dual Tumor Suppressor and Tumor Promoter Action of Sirtuins in Determining Malignant Phenotype. Front. Pharm. 2019, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Carafa, V.; Rotili, D.; Forgione, M.; Cuomo, F.; Serretiello, E.; Hailu, G.S.; Jarho, E.; Lahtela-Kakkonen, M.; Mai, A.; Altucci, L. Sirtuin functions and modulation: From chemistry to the clinic. Clin. Epigenet. 2016, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Bitterman, K.J.; Anderson, R.M.; Cohen, H.Y.; Latorre-Esteves, M.; Sinclair, D.A. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 2002, 277, 45099–45107. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, J.; Chen, D.; Yan, L.; Zheng, W. Sirtuin Inhibition: Strategies, Inhibitors, and Therapeutic Potential. Trends Pharm. Sci. 2017, 38, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Villalba, J.M.; Alcain, F.J. Sirtuin activators and inhibitors. Biofactors 2012, 38, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Gertz, M.; Fischer, F.; Nguyen, G.T.; Lakshminarasimhan, M.; Schutkowski, M.; Weyand, M.; Steegborn, C. Ex-527 inhibits Sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, E2772–E2781. [Google Scholar] [CrossRef] [PubMed]
- Napper, A.D.; Hixon, J.; McDonagh, T.; Keavey, K.; Pons, J.F.; Barker, J.; Yau, W.T.; Amouzegh, P.; Flegg, A.; Hamelin, E.; et al. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J. Med. Chem. 2005, 48, 8045–8054. [Google Scholar] [CrossRef] [PubMed]
- Botta, L.; Filippi, S.; Bizzarri, B.M.; Meschini, R.; Caputo, M.; Proietti-De-Santis, L.; Iside, C.; Nebbioso, A.; Gualandi, G.; Saladino, R. Oxidative nucleophilic substitution selectively produces cambinol derivatives with antiproliferative activity on bladder cancer cell lines. Bioorg. Med. Chem. Lett. 2019, 29, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Heltweg, B.; Gatbonton, T.; Schuler, A.D.; Posakony, J.; Li, H.; Goehle, S.; Kollipara, R.; Depinho, R.A.; Gu, Y.; Simon, J.A.; et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006, 66, 4368–4377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lara, E.; Mai, A.; Calvanese, V.; Altucci, L.; Lopez-Nieva, P.; Martinez-Chantar, M.L.; Varela-Rey, M.; Rotili, D.; Nebbioso, A.; Ropero, S.; et al. Salermide, a Sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene 2009, 28, 781–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotili, D.; Tarantino, D.; Nebbioso, A.; Paolini, C.; Huidobro, C.; Lara, E.; Mellini, P.; Lenoci, A.; Pezzi, R.; Botta, G.; et al. Discovery of salermide-related sirtuin inhibitors: Binding mode studies and antiproliferative effects in cancer cells including cancer stem cells. J. Med. Chem. 2012, 55, 10937–10947. [Google Scholar] [CrossRef] [PubMed]
- Nebbioso, A.; Pereira, R.; Khanwalkar, H.; Matarese, F.; Garcia-Rodriguez, J.; Miceli, M.; Logie, C.; Kedinger, V.; Ferrara, F.; Stunnenberg, H.G.; et al. Death receptor pathway activation and increase of ROS production by the triple epigenetic inhibitor UVI5008. Mol. Cancer 2011, 10, 2394–2404. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.; Benedetti, R.; Perez-Rodriguez, S.; Nebbioso, A.; Garcia-Rodriguez, J.; Carafa, V.; Stuhldreier, M.; Conte, M.; Rodriguez-Barrios, F.; Stunnenberg, H.G.; et al. Indole-derived psammaplin A analogues as epigenetic modulators with multiple inhibitory activities. J. Med. Chem. 2012, 55, 9467–9491. [Google Scholar] [CrossRef] [PubMed]
- Lain, S.; Hollick, J.J.; Campbell, J.; Staples, O.D.; Higgins, M.; Aoubala, M.; McCarthy, A.; Appleyard, V.; Murray, K.E.; Baker, L.; et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell 2008, 13, 454–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Maruwge, W.; Strambi, A.; D’Arcy, P.; Pellegrini, P.; Kis, L.; de Milito, A.; Lain, S.; Brodin, B. SIRT1 and SIRT2 inhibition impairs pediatric soft tissue sarcoma growth. Cell Death Dis. 2014, 5, e1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carafa, V.; Nebbioso, A.; Cuomo, F.; Rotili, D.; Cobellis, G.; Bontempo, P.; Baldi, A.; Spugnini, E.P.; Citro, G.; Chambery, A.; et al. RIP1-HAT1-SIRT Complex Identification and Targeting in Treatment and Prevention of Cancer. Clin. Cancer Res. 2018, 24, 2886–2900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mai, A.; Valente, S.; Meade, S.; Carafa, V.; Tardugno, M.; Nebbioso, A.; Galmozzi, A.; Mitro, N.; De Fabiani, E.; Altucci, L.; et al. Study of 1,4-dihydropyridine structural scaffold: Discovery of novel sirtuin activators and inhibitors. J. Med. Chem. 2009, 52, 5496–5504. [Google Scholar] [CrossRef] [PubMed]
- Spallotta, F.; Cencioni, C.; Straino, S.; Nanni, S.; Rosati, J.; Artuso, S.; Manni, I.; Colussi, C.; Piaggio, G.; Martelli, F.; et al. A nitric oxide-dependent cross-talk between class I and III histone deacetylases accelerates skin repair. J. Biol. Chem. 2013, 288, 11004–11012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valente, S.; Mellini, P.; Spallotta, F.; Carafa, V.; Nebbioso, A.; Polletta, L.; Carnevale, I.; Saladini, S.; Trisciuoglio, D.; Gabellini, C.; et al. 1,4-Dihydropyridines Active on the SIRT1/AMPK Pathway Ameliorate Skin Repair and Mitochondrial Function and Exhibit Inhibition of Proliferation in Cancer Cells. J. Med. Chem. 2016, 59, 1471–1491. [Google Scholar] [CrossRef] [PubMed]
- Sonnemann, J.; Marx, C.; Becker, S.; Wittig, S.; Palani, C.D.; Kramer, O.H.; Beck, J.F. p53-dependent and p53-independent anticancer effects of different histone deacetylase inhibitors. Br. J. Cancer 2014, 110, 656–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Yang, H.; Kong, Q.; Li, J.; Lee, S.M.; Gao, B.; Dong, H.; Wei, J.; Song, J.; Zhang, D.D.; et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell 2012, 46, 484–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosch-Presegue, L.; Vaquero, A. The dual role of sirtuins in cancer. Genes Cancer 2011, 2, 648–662. [Google Scholar] [CrossRef] [PubMed]
- Park, E.Y.; Woo, Y.; Kim, S.J.; Kim, D.H.; Lee, E.K.; De, U.; Kim, K.S.; Lee, J.; Jung, J.H.; Ha, K.T.; et al. Anticancer Effects of a New SIRT Inhibitor, MHY2256, against Human Breast Cancer MCF-7 Cells via Regulation of MDM2-p53 Binding. Int. J. Biol. Sci. 2016, 12, 1555–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.W.; Kim, S.A.; Ahn, S.G. Sirtuin inhibitors, EX527 and AGK2, suppress cell migration by inhibiting HSF1 protein stability. Oncol. Rep. 2016, 35, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Castoria, G.; Giovannelli, P.; Di Donato, M.; Hayashi, R.; Arra, C.; Appella, E.; Auricchio, F.; Migliaccio, A. Targeting androgen receptor/Src complex impairs the aggressive phenotype of human fibrosarcoma cells. PLoS ONE 2013, 8, e76899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giovannelli, P.; Di Donato, M.; Auricchio, F.; Castoria, G.; Migliaccio, A. Androgens Induce Invasiveness of Triple Negative Breast Cancer Cells Through AR/Src/PI3-K Complex Assembly. Sci. Rep. 2019, 9, 4490. [Google Scholar] [CrossRef] [PubMed] [Green Version]









 | ||||
| Compd | R1 | R2 | R3 | R4 |
| MC2494 |  | H | CN |  |
| 1 |  | CH3 | CN |  |
| 2 | H | H | CN |  |
| 3 | H | CH3 | CN |  |
| 4 |  | H | CN |  |
| 5 |  | H | CN |  |
| 6 |  | H | CN |  |
| 7 |  | H | CN |  |
| 8 |  | H | CN |  |
| 9 |  | H | CN |  |
| 10 |  | H | CN |  |
| 11 |  | H | CN |  |
| 12 |  | H | CN |  |
| 13 |  | H | CN |  |
| 14 |  | H | CN |  |
| 15 |  | H | CN |  |
| 16 |  | H | COOEt |  |
| 17 |  | H | COOEt |  |
| 18 |  | H | COOEt |  |
| 19 |  | H | COOEt |  |
| 20 |  | H | COOEt |  |
| 21 |  | H | COOEt |  |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carafa, V.; Poziello, A.; Della Torre, L.; Giovannelli, P.; Di Donato, M.; Safadeh, E.; Yu, Z.; Baldi, A.; Castoria, G.; Tomaselli, D.; et al. Enzymatic and Biological Characterization of Novel Sirtuin Modulators against Cancer. Int. J. Mol. Sci. 2019, 20, 5654. https://doi.org/10.3390/ijms20225654
Carafa V, Poziello A, Della Torre L, Giovannelli P, Di Donato M, Safadeh E, Yu Z, Baldi A, Castoria G, Tomaselli D, et al. Enzymatic and Biological Characterization of Novel Sirtuin Modulators against Cancer. International Journal of Molecular Sciences. 2019; 20(22):5654. https://doi.org/10.3390/ijms20225654
Chicago/Turabian StyleCarafa, Vincenzo, Angelita Poziello, Laura Della Torre, Pia Giovannelli, Marzia Di Donato, Elham Safadeh, Zhijun Yu, Alfonso Baldi, Gabriella Castoria, Daniela Tomaselli, and et al. 2019. "Enzymatic and Biological Characterization of Novel Sirtuin Modulators against Cancer" International Journal of Molecular Sciences 20, no. 22: 5654. https://doi.org/10.3390/ijms20225654
APA StyleCarafa, V., Poziello, A., Della Torre, L., Giovannelli, P., Di Donato, M., Safadeh, E., Yu, Z., Baldi, A., Castoria, G., Tomaselli, D., Mai, A., Rotili, D., Nebbioso, A., & Altucci, L. (2019). Enzymatic and Biological Characterization of Novel Sirtuin Modulators against Cancer. International Journal of Molecular Sciences, 20(22), 5654. https://doi.org/10.3390/ijms20225654