Profiling of LINE-1-Related Genes in Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Long Interspersed Element 1 (L1)
3. L1-Related Genes in Hepatocellular Carcinoma (HCC)
3.1. Host Defense Genes Against L1
3.2. L1-Related DDR Genes
3.3. L1 de novo Insertions
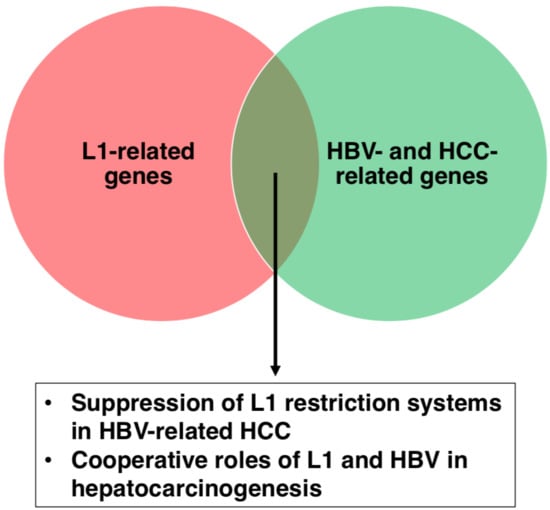
4. HBV- and HCC-Related Genes in L1 Biology
4.1. Myc
4.2. CBX1, Rad21 and CENPA
4.3. HBV Insertions
4.4. The HBx-L1 Chimeric Transcript
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ferenci, P.; Fried, M.; Labrecque, D.; Bruix, J.; Sherman, M.; Omata, M.; Heathcote, J.; Piratsivuth, T.; Kew, M.; Otegbayo, J.A.; et al. Hepatocellular Carcinoma (HCC). J. Clin. Gastroenterol. 2010, 44, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer Incidence and Mortality Rates and Trends—An Update. Cancer Epidemiol. Biomark. Prev. 2016, 25, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Hollinger, F.B.; Liang, T.J. Hepatitis B Virus, 4th ed.; Fields Virology; Lippincott-Raven Publishers: Philadelphia, PA, USA, 2001. [Google Scholar]
- Ghouri, Y.A.; Mian, I.; Rowe, J.H. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J. Carcinog. 2017, 16, 1. [Google Scholar] [PubMed]
- Ganem, D.; Prince, A.M. Hepatitis B Virus Infection—Natural History and Clinical Consequences. N. Engl. J. Med. 2004, 350, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Kim, W.R.; Coelho, R.; Mettler, T.A.; Benson, J.T.; Sanderson, S.O.; Therneau, T.M.; Kim, B.; Roberts, L.R. Cirrhosis is Present in Most Patients With Hepatitis B and Hepatocellular Carcinoma. Clin. Gastroenterol. Hepatol. 2011, 9, 64–70. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Hepatitis B World Health Organization Fact Sheet. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b (accessed on 30 December 2018).
- Li, X.; Zhao, J.; Yuan, Q.; Xia, N. Detection of HBV Covalently Closed Circular DNA. Viruses 2017, 9, 139. [Google Scholar] [CrossRef] [PubMed]
- Summers, J.; O’Connell, A.; Millman, I. Genome of hepatitis B virus: Restriction enzyme cleavage and structure of DNA extracted from Dane particles. Proc. Natl. Acad. Sci. USA 1975, 72, 4597–4601. [Google Scholar] [CrossRef]
- Beck, J.; Nassal, M. Hepatitis B virus replication. World J. Gastroenterol. 2007, 13, 48–64. [Google Scholar] [CrossRef]
- Seeger, C.; Mason, W.S. Hepatitis B Virus Biology. Microbiol. Mol. Biol. Rev. 2000, 64, 51–68. [Google Scholar] [CrossRef] [Green Version]
- Patient, R.; Hourioux, C.; Sizaret, P.Y.; Trassard, S.; Sureau, C.; Roingeard, P. Hepatitis B virus subviral envelope particle morphogenesis and intracellular trafficking. J. Virol. 2007, 81, 3842–3851. [Google Scholar] [CrossRef]
- Tang, Z.Y. Hepatocellular carcinoma-cause, treatment and metastasis. World J. Gastroenterol. 2001, 7, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Benvegnù, L.; Gios, M.; Boccato, S.; Alberti, A. Natural history of compensated viral cirrhosis: A prospective study on the incidence and hierarchy of major complications. Gut 2004, 53, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Petersen, G.M. Familial Aggregation: Sorting Susceptibility From Shared Environment. J. Natl. Cancer Inst. 2000, 92, 1114–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohnishi, K.; Iida, S.; Iwama, S.; Goto, N.; Nomura, F.; Takashi, M.; Mishima, A.; Kono, K.; Kimura, K.; Musha, H.; et al. The Effect of Chronic Habitual Alcohol Intake on the Development of Liver Cirrhosis and Hepatocellular Carcinoma: Relation to Hepatitis B Surface Antigen Carriage. Am. Cancer Soc. 1981, 672–677. [Google Scholar] [CrossRef]
- Bae, S.H.; Yoon, S.K.; Jang, J.W.; Kim, C.W.; Nam, S.W.; Choi, J.Y.; Kim, B.S.; Park, Y.M.; Suzuki, S.; Sugauchi, F.; et al. Hepatitis B virus genotype C prevails among chronic carriers of the virus in Korea. J. Korean Med. Sci. 2005, 20, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.W.; Yeh, S.H.H.; Chen, P.J.J.; Liaw, Y.F.F.; Lin, C.L.L.; Liu, C.J.J.; Shih, W.L.L.; Kao, J.H.H.; Chen, D.S.S.; Chen, C.J.J. Hepatitis B Virus Genotype and DNA Level and Hepatocellular Carcinoma: A Prospective Study in Men. J. Natl. Cancer Inst. 2005, 97, 265–272. [Google Scholar] [CrossRef] [Green Version]
- Kao, J.; Chen, P.; Lai, M.; Chen, D. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology 2003, 124, 327–334. [Google Scholar] [CrossRef]
- Yang, H.I.; Lu, S.N.; Liaw, Y.F.; You, S.L.; Sun, C.A.; Wang, L.Y.; Hsiao, C.K.; Chen, P.J.; Chen, D.S.; Chen, C.J.; et al. Hepatitis B e Antigen and the Risk of Hepatocellular Carcinoma. N. Engl. J. Med. 2002, 347, 168–174. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar]
- Beck, C.R.; Collier, P.; Macfarlane, C.; Malig, M.; Kidd, J.M.; Eichler, E.E.; Badge, R.M.; Moran, J.V. LINE-1 retrotransposition activity in human genomes. Cell 2010, 141, 1159–1170. [Google Scholar] [CrossRef]
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Moran, J.V.; Kazazian, H.H. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 2003, 100, 5280–5285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, K.H.; Boeke, J.D. Human Transposon Tectonics. Cell 2012, 149, 740–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, H.L.; Moran, J.V. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 2011, 12, 615–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodić, N.; Sharma, R.; Sharma, R.; Zampella, J.; Dai, L.; Taylor, M.S.; Hruban, R.H.; Iacobuzio-Donahue, C.A.; Maitra, A.; Torbenson, M.S.; et al. Long Interspersed Element-1 Protein Expression Is a Hallmark of Many Human Cancers. Am. J. Pathol. 2014, 184, 1280–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangasamy, D.; Lenka, N.; Ohms, S.; Dahlstrom, J.E.; Blackburn, A.C.; Board, P.G. Activation of LINE-1 Retrotransposon Increases the Risk of Epithelial-Mesenchymal Transition and Metastasis in Epithelial Cancer. Curr. Mol. Med. 2015, 15, 588–597. [Google Scholar] [PubMed]
- Kemp, J.R.; Longworth, M.S. Crossing the LINE Toward Genomic Instability: LINE-1 Retrotransposition in Cancer. Front. Chem. 2015, 3, 68. [Google Scholar] [CrossRef] [PubMed]
- Briggs, E.M.; Ha, S.; Mita, P.; Brittingham, G.; Sciamanna, I.; Spadafora, C.; Logan, S.K. Long interspersed nuclear element-1 expression and retrotransposition in prostate cancer cells. Mob. DNA 2018, 9, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.; Iskow, R.; Yang, L.; Gokcumen, O.; Haseley, P.; Luquette, L.J.; Lohr, J.G.; Harris, C.C.; Ding, L.; Wilson, R.K.; et al. Landscape of somatic retrotransposition in human cancers. Science 2012, 337, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Upton, K.R.; Muñoz-Lopez, M.; Gerhardt, D.J.; Fisher, M.E.; Nguyen, T.; Brennan, P.M.; Baillie, J.K.; Collino, A.; Ghisletti, S.; et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 2013, 153, 101–111. [Google Scholar] [CrossRef]
- Honda, T. Links between Human LINE-1 Retrotransposons and Hepatitis Virus-Related Hepatocellular Carcinoma. Front. Chem. 2016, 4, 21. [Google Scholar] [CrossRef]
- Honda, T. Potential Links between Hepadnavirus and Bornavirus Sequences in the Host Genome and Cancer. Front. Microbiol. 2017, 8, 2537. [Google Scholar] [CrossRef] [PubMed]
- Schauer, S.N.; Carreira, P.E.; Shukla, R.; Gerhardt, D.J.; Gerdes, P.; Sanchez-Luque, F.J.; Nicoli, P.; Kindlova, M.; Ghisletti, S.; Dos Santos, A.; et al. L1 retrotransposition is a common feature of mammalian hepatocarcinogenesis. Genome Res. 2018, 28, 639–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodier, J.L. Retrotransposition in tumors and brains. Mob. DNA 2014, 5, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, C.V.; Asch, A.S.; Yamada, H.Y. Frequently mutated genes/pathways and genomic instability as prevention targets in liver cancer. Carcinogenesis 2017, 38, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.C.C.; Mayakonda, A.; Dinh, H.Q.; Huang, P.; Lin, L.; Liu, X.; Ding, L.W.; Wang, J.; Berman, B.P.; Song, E.W.W.; et al. Genomic and Epigenomic Heterogeneity of Hepatocellular Carcinoma. Cancer Res. 2017, 77, 2255–2265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, C.C.; Sun, T.; Ching, A.K.K.; He, M.; Li, J.W.; Wong, A.M.; Co, N.N.; Chan, A.W.H.; Li, P.S.; Lung, R.W.M.; et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell 2014, 25, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Khazina, E.; Truffault, V.; Büttner, R.; Schmidt, S.; Coles, M.; Weichenrieder, O. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol. 2011, 18, 1006–1014. [Google Scholar] [CrossRef]
- Feng, Q.; Moran, J.V.; Kazazian, H.H.; Boeke, J.D. Human L1 Retrotransposon Encodes a Conserved Endonuclease Required for Retrotransposition. Cell 1996, 87, 905–916. [Google Scholar] [CrossRef]
- Terasaki, N.; Goodier, J.L.; Cheung, L.E.; Wang, Y.J.; Kajikawa, M.; Kazazian, H.H.; Okada, N. In vitro screening for compounds that enhance human L1 mobilization. PLoS ONE 2013, 8, e74629. [Google Scholar] [CrossRef]
- Giorgi, G.; Marcantonio, P.; Del Re, B. LINE-1 retrotransposition in human neuroblastoma cells is affected by oxidative stress. Cell Tissue Res. 2011, 346, 383–391. [Google Scholar] [CrossRef]
- Bundo, M.; Toyoshima, M.; Okada, Y.; Akamatsu, W.; Ueda, J.; Nemoto-Miyauchi, T.; Sunaga, F.; Toritsuka, M.; Ikawa, D.; Kakita, A.; et al. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron 2014, 81, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.B.; Song, H.; Xu, Y.; Garrison, K.E.; Buzdin, A.A.; Anwar, N.; Hunter, D.V.; Mujib, S.; Mihajlovic, V.; Martin, E.; et al. LINE-1 retrotransposable element DNA accumulates in HIV-1-infected cells. J. Virol. 2013, 87, 13307–13320. [Google Scholar] [CrossRef] [PubMed]
- Iijima, K.; Okudaira, N.; Tamura, M.; Doi, A.; Saito, Y.; Shimura, M.; Goto, M.; Matsunaga, A.; Kawamura, Y.I.; Otsubo, T.; et al. Viral protein R of human immunodeficiency virus type-1 induces retrotransposition of long interspersed element-1. Retrovirology 2013, 10, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burwinkel, B.; Kilimann, M.W. Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J. Mol. Biol. 1998, 277, 513–517. [Google Scholar] [CrossRef]
- Mbisa, J.L.; Bu, W.; Pathak, V.K. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J. Virol. 2010, 84, 5250–5259. [Google Scholar] [CrossRef] [PubMed]
- Nowarski, R.; Prabhu, P.; Kenig, E.; Smith, Y.; Britan-Rosich, E.; Kotler, M. APOBEC3G Inhibits HIV-1 RNA Elongation by Inactivating the Viral Trans-Activation Response Element. J. Mol. Biol. 2014, 426, 2840–2853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinomoto, M.; Kanno, T.; Shimura, M.; Ishizaka, Y.; Kojima, A.; Kurata, T.; Sata, T.; Tokunaga, K. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007, 35, 2955–2964. [Google Scholar] [CrossRef] [Green Version]
- Antonucci, J.M.; St. Gelais, C.; Wu, L. The Dynamic Interplay between HIV-1, SAMHD1, and the Innate Antiviral Response. Front. Immunol. 2017, 8, 1541. [Google Scholar] [CrossRef]
- Hu, S.; Li, J.; Xu, F.; Mei, S.; Le Duff, Y.; Yin, L.; Pang, X.; Cen, S.; Jin, Q.; Liang, C.; et al. SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation. PLoS Genet. 2015, 11, e1005367. [Google Scholar] [CrossRef]
- Burdick, R.; Smith, J.L.; Chaipan, C.; Friew, Y.; Chen, J.; Venkatachari, N.J.; Delviks-Frankenberry, K.A.; Hu, W.-S.; Pathak, V.K. P Body-Associated Protein Mov10 Inhibits HIV-1 Replication at Multiple Stages. J. Virol. 2010, 84, 10241–10253. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhang, J.; Jia, R.; Cheng, V.; Xu, X.; Qiao, W.; Guo, F.; Liang, C.; Cen, S. The MOV10 Helicase Inhibits LINE-1 Mobility. J. Biol. Chem. 2013, 288, 21148–21160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janahi, E.M.; McGarvey, M.J. The inhibition of hepatitis B virus by APOBEC cytidine deaminases. J. Viral Hepat. 2013, 20, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Mohamadkhani, A.; Pourdadash, A.; Tayebi, S.; Estakhri, A.; Nazem, H.; Sotoudeh, M.; Poustchi, H. The potential role of APOBEC3G in limiting replication of hepatitis B virus. Arab J. Gastroenterol. 2012, 13, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Wang, X.; Lou, G.; Peng, G.; Li, J.; Zhu, H.; Chen, F.; Li, S.; Liu, D.; Chen, Z.; et al. APOBEC3G directly binds Hepatitis B virus core protein in cell and cell free systems. Virus Res. 2010, 151, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Turelli, P.; Mangeat, B.; Jost, S.; Vianin, S.; Trono, D. Inhibition of Hepatitis B Virus Replication by APOBEC3G. Science 2004, 303, 1829. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Wang, C.; Ma, X.; Wang, E.; Peng, G. APOBEC3B, a molecular driver of mutagenesis in human cancers. Cell Biosci. 2017, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Zhang, X.; Zhang, W.; Fang, Y.; Zheng, S.; Yu, X.F. Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatology 2007, 46, 1810–1820. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Zhu, M.; Pan, X.; Zhu, Y.; Yan, H.; Jiang, T.; Shen, Y.Y.; Dong, X.; Zheng, N.; Lu, J.; et al. Inhibition of Hepatitis B virus replication by SAMHD1. Biochem. Biophys. Res. Commun. 2014, 450, 1462–1468. [Google Scholar] [CrossRef]
- Shi, Y.; Lv, G.; Chu, Z.; Piao, L.; Liu, X.; Wang, T.; Jiang, Y.; Zhang, P. Identification of natural splice variants of SAMHD1 in virus-infected HCC. Oncol. Rep. 2014, 31, 687–692. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, L.; Ying, S. SAMHD1: A novel antiviral factor in intrinsic immunity. Future Microbiol. 2012, 7, 1117–1126. [Google Scholar] [CrossRef]
- Hollenbaugh, J.A.; Gee, P.; Baker, J.; Daly, M.B.; Amie, S.M.; Tate, J.; Kasai, N.; Kanemura, Y.; Kim, D.H.; Ward, B.M.; et al. Host Factor SAMHD1 Restricts DNA Viruses in Non-Dividing Myeloid Cells. PLoS Pathog. 2013, 9, e1003481. [Google Scholar] [CrossRef] [PubMed]
- Gramberg, T.; Kahle, T.; Bloch, N.; Wittmann, S.; Müllers, E.; Daddacha, W.; Hofmann, H.; Kim, B.; Lindemann, D.; Landau, N.R. Restriction of diverse retroviruses by SAMHD1. Retrovirology 2013, 10, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldstone, D.C.; Ennis-Adeniran, V.; Hedden, J.J.; Groom, H.C.T.; Rice, G.I.; Christodoulou, E.; Walker, P.A.; Kelly, G.; Haire, L.F.; Yap, M.W.; et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 2011, 480, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Laguette, N.; Sobhian, B.; Casartelli, N.; Ringeard, M.; Chable-Bessia, C.; Ségéral, E.; Yatim, A.; Emiliani, S.; Schwartz, O.; Benkirane, M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 2011, 474, 654–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahouassa, H.; Daddacha, W.; Hofmann, H.; Ayinde, D.; Logue, E.C.; Dragin, L.; Bloch, N.; Maudet, C.; Bertrand, M.; Gramberg, T.; et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 2012, 13, 223–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.; Ryoo, J.; Oh, C.; Hwang, S.; Ahn, K. SAMHD1 specifically restricts retroviruses through its RNase activity. Retrovirology 2015, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Du, J.; Han, X.; Goodier, J.L.; Li, P.; Zhou, X.; Wei, W.; Evans, S.L.; Li, L.; Zhang, W.; et al. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutières syndrome-related SAMHD1. Cell Rep. 2013, 4, 1108–1115. [Google Scholar] [CrossRef] [PubMed]
- Jeong, G.U.; Park, I.H.; Ahn, K.; Ahn, B.Y. Inhibition of hepatitis B virus replication by a dNTPase-dependent function of the host restriction factor SAMHD1. Virology 2016, 495, 71–78. [Google Scholar] [CrossRef]
- Welbourn, S.; Miyagi, E.; White, T.E.; Diaz-Griffero, F.; Strebel, K. Identification and characterization of naturally occurring splice variants of SAMHD1. Retrovirology 2012, 9, 86. [Google Scholar] [CrossRef]
- Furtak, V.; Mulky, A.; Rawlings, S.A.; Kozhaya, L.; Lee, K.; KewalRamani, V.N.; Unutmaz, D. Perturbation of the P-Body Component Mov10 Inhibits HIV-1 Infectivity. PLoS ONE 2010, 5, e9081. [Google Scholar] [CrossRef]
- Wang, X.; Han, Y.; Dang, Y.; Fu, W.; Zhou, T.; Ptak, R.G.; Zheng, Y.H. Moloney Leukemia Virus 10 (MOV10) Protein Inhibits Retrovirus Replication. J. Biol. Chem. 2010, 285, 14346–14355. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.X.; Li, D.; Fu, L.J.; Fu, B.Q.; Chen, S.J.; Xu, W.Z.; Teng, X.; Song, Z.W.; Gu, H.X. The role of Moloney leukemia virus 10 in hepatitis B virus expression in hepatoma cells. Virus Res. 2015, 197, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.W.; Ma, Y.X.; Fu, B.Q; Teng, X.; Chen, S.J.; Xu, W.Z.; Gu, H.X. Altered mRNA levels of MOV10, A3G, and IFN-α in patients with chronic hepatitis B. J. Microbiol. 2014, 52, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Ditch, S.; Paull, T.T. The ATM protein kinase and cellular redox signaling: Beyond the DNA damage response. Trends Biochem. Sci. 2012, 37, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Tho, L.M.; Xu, N.; Gillespie, D.A. The ATM–Chk2 and ATR–Chk1 Pathways in DNA Damage Signaling and Cancer. Adv Cancer Res. 2010, 108, 73–112. [Google Scholar] [PubMed]
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Marchetto, M.C.N.; Muotri, A.R.; Mu, Y.; Carson, C.T.; Macia, A.; Moran, J.V.; Gage, F.H. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl. Acad. Sci. USA 2011, 108, 20382–20387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Lee, H.S.; Ji, J.H.; Cho, M.Y.; Yoo, Y.S.; Park, Y.Y.; Cha, H.J.; Lee, Y.; Kim, Y.; Cho, H. Hepatitis B virus X protein activates the ATM-Chk2 pathway and delays cell cycle progression. J. Gen. Virol. 2015, 96, 2242–2251. [Google Scholar] [CrossRef]
- Zhao, F.; Hou, N.B.; Song, T.; He, X.; Zheng, Z.R.; Ma, Q.J.; Li, L.; Zhang, Y.H.; Zhong, H. Cellular DNA repair cofactors affecting hepatitis B virus infection and replication. World J. Gastroenterol. 2008, 14, 5059–5065. [Google Scholar] [CrossRef]
- Sen, S.K.; Huang, C.T.; Han, K.; Batzer, M.A. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Res. 2007, 35, 3741–3751. [Google Scholar] [CrossRef] [Green Version]
- Dumble, M.L.; Croager, E.J.; Yeoh, G.C.T.; Quail, E.A. Generation and characterization of p53 null transformed hepatic progenitor cells: Oval cells give rise to hepatocellular carcinoma. Carcinogenesis 2002, 23, 435–445. [Google Scholar] [CrossRef]
- Lechel, A.; Holstege, H.; Begus, Y.; Schienke, A.; Kamino, K.; Lehmann, U.; Kubicka, S.; Schirmacher, P.; Jonkers, J.; Rudolph, K.L. Telomerase Deletion Limits Progression of p53-Mutant Hepatocellular Carcinoma With Short Telomeres in Chronic Liver Disease. Gastroenterology 2007, 132, 1465–1475. [Google Scholar] [CrossRef] [PubMed]
- Kunst, C.; Haderer, M.; Heckel, S.; Schlosser, S.; Müller, M. The p53 family in hepatocellular carcinoma. Transl. Cancer Res. 2016, 5, 632–638. [Google Scholar] [CrossRef]
- Wylie, A.; Jones, A.E.; D’Brot, A.; Lu, W.J.; Kurtz, P.; Moran, J.V.; Rakheja, D.; Chen, K.S.; Hammer, R.E.; Comerford, S.A.; et al. p53 genes function to restrain mobile elements. Genes Dev. 2016, 30, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Shiraha, H.; Yamamoto, K.; Namba, M. Human hepatocyte carcinogenesis. Int. J. Oncol. 2013, 42, 1133–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Franklin, D.A.; Dong, J.; Zhang, Y. MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res. 2014, 74, 7161–7167. [Google Scholar] [CrossRef]
- Soini, Y.; Chia, S.C.; Bennett, W.P.; Groopman, J.D.; Wang, J.S.; DeBenedetti, V.M.; Cawley, H.; Welsh, J.A.; Hansen, C.; Bergasa, N.V.; et al. An aflatoxin-associated mutational hotspot at codon 249 in the p53 tumor suppressor gene occurs in hepatocellular carcinomas from Mexico. Carcinogenesis 1996, 17, 1007–1012. [Google Scholar] [CrossRef] [Green Version]
- Stähler, F.; Roemer, K. Mutant p53 can provoke apoptosis in p53-deficient Hep3B cells with delayed kinetics relative to wild-type p53. Oncogene 1998, 17, 3507–3512. [Google Scholar] [CrossRef] [Green Version]
- Ueda, H.; Ullrich, S.J.; Gangemi, J.D.; Kappel, C.A.; Ngo, L.; Feitelson, M.A.; Jay, G. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat. Genet. 1995, 9, 41–47. [Google Scholar] [CrossRef]
- Anzola, M.; Cuevas, N.; López-Martínez, M.; Saiz, A.; Burgos, J.J.; Martínez de Pancorboa, M. p14ARF gene alterations in human hepatocellular carcinoma. Eur. J. Gastroenterol. Hepatol. 2004, 16, 19–26. [Google Scholar] [CrossRef]
- Wade, M.; Li, Y.C.; Wahl, G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 2013, 13, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Feitelson, M.A.; Zhu, M.; Duan, L.X.; London, W.T. Hepatitis B x antigen and p53 are associated in vitro and in liver tissues from patients with primary hepatocellular carcinoma. Oncogene 1993, 8, 1109–1117. [Google Scholar] [PubMed]
- Zhu, M.; London, W.T.; Duan, L.X.; Feitelson, M.A. The value of hepatitis B x antigen as a prognostic marker in the development of hepatocellular carcinoma. Int. J. Cancer 1993, 55, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Feitelson, M.A.; Ranganathan, P.N.; Clayton, M.M.; Zhang, S.M. Partial characterization of the woodchuck tumor suppressor, p53, and its interaction with woodchuck hepatitis virus X antigen in hepatocarcinogenesis. Oncogene 1997, 15, 327–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuyama, R.; Niculaita, R.; Ng, K.P.; Obusez, E.; Sanchez, J.; Kalady, M.; Aung, P.P.; Casey, G.; Sizemore, N. Mutated in colorectal cancer, a putative tumor suppressor for serrated colorectal cancer, selectively represses beta-catenin-dependent transcription. Oncogene 2008, 27, 6044–6055. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Qu, J.; Chang, X.; Lu, Y.; Bai, W.; Wang, H.; Xu, Z.; An, L.; Wang, C.; Zeng, Z.; et al. Hypomethylation of long interspersed nuclear element-1 promoter is associated with poor outcomes for curative resected hepatocellular carcinoma. Liver Int. 2014, 34, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Utsunomiya, T.; Ikemoto, T.; Yamada, S.; Morine, Y.; Imura, S.; Arakawa, Y.; Takasu, C.; Ishikawa, D.; Imoto, I.; et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) is associated with poor prognosis via activation of c-MET in hepatocellular carcinoma. Ann. Surg. Oncol. 2014, 21 (Suppl. 4), S729–S735. [Google Scholar] [CrossRef]
- Zhang, C.; Fan, L.; Fan, T.; Wu, D.; Gao, L.; Ling, Y.; Zhu, J.; Li, R.; Wei, L. Decreased PADI4 mRNA association with global hypomethylation in hepatocellular carcinoma during HBV exposure. Cell Biochem. Biophys. 2013, 65, 187–195. [Google Scholar] [CrossRef]
- Shitani, M.; Sasaki, S.; Akutsu, N.; Takagi, H.; Suzuki, H.; Nojima, M.; Yamamoto, H.; Tokino, T.; Hirata, K.; Imai, K.; et al. Genome-wide analysis of DNA methylation identifies novel cancer-related genes in hepatocellular carcinoma. Tumour Biol. 2012, 33, 1307–1317. [Google Scholar] [CrossRef]
- Lee, W.Y.; Bachtiar, M.; Choo, C.C.S.; Lee, C.G. Comprehensive review of Hepatitis B Virus-associated hepatocellular carcinoma research through text mining and big data analytics. Biol. Rev. 2018. [Google Scholar] [CrossRef]
- Lin, C.P.; Liu, C.R.; Lee, C.N.; Chan, T.S.; Liu, H.E. Targeting c-Myc as a novel approach for hepatocellular carcinoma. World J. Hepatol. 2010, 2, 16–20. [Google Scholar] [CrossRef]
- Balsano, C.; Avantaggiati, M.L.; Natoli, G.; De Marzio, E.; Will, H.; Perricaudet, M.; Levrero, M. Full-length and truncated versions of the hepatitis B virus (HBV) X protein (pX) transactivate the cmyc protooncogene at the transcriptional level. Biochem. Biophys. Res. Commun. 1991, 176, 985–992. [Google Scholar] [CrossRef]
- Lee, S.; Kim, W.; Ko, C.; Ryu, W.S. Hepatitis B virus X protein enhances Myc stability by inhibiting SCFSkp2 ubiquitin E3 ligase-mediated Myc ubiquitination and contributes to oncogenesis. Oncogene 2016, 35, 1857–1867. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, N.; Tsunedomi, R.; Tamesa, T.; Okada, T.; Sakamoto, K.; Hamaguchi, T.; Yamada-Okabe, H.; Miyamoto, T.; Uchimura, S.; Hamamoto, Y.; et al. Involvement of c-myc-regulated genes in hepatocellular carcinoma related to genotype-C hepatitis B virus. J. Cancer Res. Clin. Oncol. 2006, 132, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Rockwood, L.D.; Felix, K.; Janz, S. Elevated presence of retrotransposons at sites of DNA double strand break repair in mouse models of metabolic oxidative stress and MYC-induced lymphoma. Mutat. Res. Mol. Mech. Mutagen. 2004, 548, 117–125. [Google Scholar] [CrossRef]
- Gasior, S.L.; Preston, G.; Hedges, D.J.; Gilbert, N.; Moran, J.V.; Deininger, P.L. Characterization of pre-insertion loci of de novo L1 insertions. Gene 2007, 390, 190–198. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.; Lin, B.; Liu, H.; Ma, X.; Mo, F.; Yu, W.; Li, L.; Li, H.; Tian, T.; Wu, D.; et al. RNA-seq analyses generate comprehensive transcriptomic landscape and reveal complex transcript patterns in hepatocellular carcinoma. PLoS ONE 2011, 6, e26168. [Google Scholar] [CrossRef]
- Boyault, S.; Rickman, D.S.; De Reyniès, A.; Balabaud, C.; Rebouissou, S.; Jeannot, E.; Hérault, A.; Saric, J.; Belghiti, J.; Franco, D.; et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 2007, 45, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Gopal, R.; Selvarasu, K.; Pandian, P.P.; Ganesan, K. Integrative transcriptome analysis of liver cancer profiles identifies upstream regulators and clinical significance of ACSM3 gene expression. Cell. Oncol. 2017, 40, 219–233. [Google Scholar] [CrossRef]
- Okabe, H.; Satoh, S.; Kato, T.; Kitahara, O.; Yanagawa, R.; Yamaoka, Y.; Tsunoda, T.; Furukawa, Y.; Nakamura, Y. Genome-wide Analysis of Gene Expression in Human Hepatocellular Carcinomas Using cDNA Microarray: Identification of Genes Involved in Viral Carcinogenesis and Tumor Progression. Cancer Res. 2001, 61, 2129–2137. [Google Scholar]
- Kaufman, P.D. New Partners for HP1 in Transcriptional Gene Silencing. Mol. Cell 2011, 41, 1–2. [Google Scholar] [CrossRef]
- Eissenberg, J.C.; Elgin, S.C. The HP1 protein family: Getting a grip on chromatin. Curr. Opin. Genet. Dev. 2000, 10, 204–210. [Google Scholar] [CrossRef]
- Yang, Y.F.; Pan, Y.H.; Tian, Q.H.; Wu, D.C.; Su, S.G. CBX1 Indicates Poor Outcomes and Exerts Oncogenic Activity in Hepatocellular Carcinoma. Transl. Oncol. 2018, 11, 1110–1118. [Google Scholar] [CrossRef] [PubMed]
- Vogel, M.J.; Guelen, L.; de Wit, E.; Hupkes, D.P.; Loden, M.; Talhout, W.; Feenstra, M.; Abbas, B.; Classen, A.K.; van Steensel, B. Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res. 2006, 16, 1493–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomonaga, T.; Nagao, K.; Kawasaki, Y.; Furuya, K.; Murakami, A.; Morishita, J.; Yuasa, T.; Sutani, T.; Kearsey, S.E.; Uhlmann, F.; et al. Characterization of fission yeast cohesin: Essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000, 14, 2757–2770. [Google Scholar] [CrossRef] [PubMed]
- Barber, T.D.; McManus, K.; Yuen, K.W.Y.; Reis, M.; Parmigiani, G.; Shen, D.; Barrett, I.; Nouhi, Y.; Spencer, F.; Markowitz, S.; et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc. Natl. Acad. Sci. USA 2008, 105, 3443–3448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Tomaszewski, J.M.; McKay, M.J. Can corruption of chromosome cohesion create a conduit to cancer? Nat. Rev. Cancer 2011, 11, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Deb, S.; Xu, H.; Tuynman, J.; George, J.; Yan, Y.; Li, J.; Ward, R.L.; Mortensen, N.; Hawkins, N.J.; McKay, M.J.; et al. RAD21 cohesin overexpression is a prognostic and predictive marker exacerbating poor prognosis in KRAS mutant colorectal carcinomas. Br. J. Cancer 2014, 110, 1606–1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Yan, Y.; Deb, S.; Rangasamy, D.; Germann, M.; Malaterre, J.; Eder, N.C.; Ward, R.L.; Hawkins, N.J.; Tothill, R.W.; et al. Cohesin Rad21 mediates loss of heterozygosity and is upregulated via Wnt promoting transcriptional dysregulation in gastrointestinal tumors. Cell Rep. 2014, 9, 1781–1797. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.W.; Craig, J.M.; Saffery, R.; Kalitsis, P.; Irvine, D.V.; Earle, E.; Magliano, D.J.; Choo, K.H. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 2001, 20, 2087–2096. [Google Scholar] [CrossRef] [Green Version]
- Amor, D.J.; Choo, K.H.A. Neocentromeres: Role in Human Disease, Evolution, and Centromere Study. Am. J. Hum. Genet. 2002, 71, 695–714. [Google Scholar] [CrossRef] [Green Version]
- Chueh, A.C.; Northrop, E.L.; Brettingham-Moore, K.H.; Choo, K.H.A.; Wong, L.H. LINE Retrotransposon RNA Is an Essential Structural and Functional Epigenetic Component of a Core Neocentromeric Chromatin. PLoS Genet. 2009, 5, e1000354. [Google Scholar] [CrossRef]
- Ding, D.; Lou, X.; Hua, D.; Yu, W.; Li, L.; Wang, J.; Gao, F.; Zhao, N.; Ren, G.; Li, L.; et al. Recurrent targeted genes of hepatitis B virus in the liver cancer genomes identified by a next-generation sequencing-based approach. PLoS Genet. 2012, 8, e1003065. [Google Scholar] [CrossRef] [PubMed]
- Pinyol, R.; Tovar, V.; Llovet, J.M. TERT promoter mutations: Gatekeeper and driver of hepatocellular carcinoma. J. Hepatol. 2014, 61, 685–687. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly Recurrent TERT Promoter Mutations in Human Melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Killela, P.J.; Reitman, Z.J.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026. [Google Scholar] [CrossRef] [Green Version]
- Nault, J.C.; Calderaro, J.; Di Tommaso, L.; Balabaud, C.; Zafrani, E.S.; Bioulac-Sage, P.; Roncalli, M.; Zucman-Rossi, J. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology 2014, 60, 1983–1992. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Guo, X.; Chen, Y.; Chen, G.; Ma, Y.; Huang, K.; Zhang, Y.; Zhao, Q.; Winkler, C.A.; An, P.; et al. Telomerase reverse transcriptase promoter mutations in hepatitis B virus-associated hepatocellular carcinoma. Oncotarget 2016, 7, 27838–27847. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Zeng, X.; Lee, N.P.; Liu, X.; Chen, S.; Guo, B.; Yi, S.; Zhuang, X.; Chen, F.; Wang, G.; et al. HIVID: An efficient method to detect HBV integration using low coverage sequencing. Genomics 2013, 102, 338–344. [Google Scholar] [CrossRef]
- Sung, W.K.; Zheng, H.; Li, S.; Chen, R.; Liu, X.; Li, Y.; Lee, N.P.; Lee, W.H.; Ariyaratne, P.N.; Tennakoon, C.; et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 2012, 44, 765–769. [Google Scholar] [CrossRef]
- Zhao, L.H.; Liu, X.; Yan, H.X.; Li, W.Y.; Zeng, X.; Yang, Y.; Zhao, J.; Liu, S.P.; Zhuang, X.H.; Lin, C.; et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat. Commun. 2016, 7, 12992. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, A.; Totoki, Y.; Abe, T.; Boroevich, K.A.; Hosoda, F.; Nguyen, H.H.; Aoki, M.; Hosono, N.; Kubo, M.; Miya, F.; et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 2012, 44, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, S.; Marais, R.; Zhu, A.X. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 2010, 29, 4989–5005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, H.W.; Wang, N.; Wang, Y.; Wang, F.; Fu, Z.; Yan, X.; Zhu, H.; Diao, W.; Ding, Y.; Chen, X.; et al. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J. Hepatol. 2016, 64, 278–291. [Google Scholar] [CrossRef] [PubMed]
- Leibold, D.M.; Swergold, G.D.; Singer, M.F.; Thayer, R.E.; Dombroski, B.A.; Fanning, T.G. Translation of LINE-1 DNA elements in vitro and in human cells. Proc. Natl. Acad. Sci. USA 1990, 87, 6990–6994. [Google Scholar] [CrossRef] [PubMed]
- Horn, A.V.; Celic, I.; Dong, C.; Martirosyan, I.; Han, J.S. A conserved role for the ESCRT membrane budding complex in LINE retrotransposition. PLoS Genet. 2017, 13, e1006837. [Google Scholar] [CrossRef] [PubMed]
- Harris, C.R.; Normart, R.; Yang, Q.; Stevenson, E.; Haffty, B.G.; Ganesan, S.; Cordon-Cardo, C.; Levine, A.J.; Tang, L.H. Association of Nuclear Localization of a Long Interspersed Nuclear Element-1 Protein in Breast Tumors with Poor Prognostic Outcomes. Genes Cancer 2010, 1, 115–124. [Google Scholar] [CrossRef]
- Chen, Y.; Zeng, Q.; Liu, X.; Fu, J.; Zeng, Z.; Zhao, Z.; Liu, Z.; Bai, W.; Dong, Z.; Liu, H.; et al. LINE-1 ORF-1p enhances the transcription factor activity of pregnenolone X receptor and promotes sorafenib resistance in hepatocellular carcinoma cells. Cancer Manag. Res. 2018, 10, 4421–4438. [Google Scholar] [CrossRef]
- Nakayama, R.; Ueno, Y.; Ueda, K.; Honda, T. Latent infection with Kaposi’s sarcoma-associated herpesvirus enhances retrotransposition of long interspersed element-1. Oncogene 2019. [Google Scholar] [CrossRef]
- Nishikawa, Y.; Nakayama, R.; Obika, S.; Ohsaki, E.; Ueda, K.; Honda, T. Inhibition of LINE-1 Retrotransposition by Capsaicin. Int. J. Mol. Sci. 2018, 19, 3243. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Honda, T.; Rahman, M.A. Profiling of LINE-1-Related Genes in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2019, 20, 645. https://doi.org/10.3390/ijms20030645
Honda T, Rahman MA. Profiling of LINE-1-Related Genes in Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2019; 20(3):645. https://doi.org/10.3390/ijms20030645
Chicago/Turabian StyleHonda, Tomoyuki, and Md. Arifur Rahman. 2019. "Profiling of LINE-1-Related Genes in Hepatocellular Carcinoma" International Journal of Molecular Sciences 20, no. 3: 645. https://doi.org/10.3390/ijms20030645
APA StyleHonda, T., & Rahman, M. A. (2019). Profiling of LINE-1-Related Genes in Hepatocellular Carcinoma. International Journal of Molecular Sciences, 20(3), 645. https://doi.org/10.3390/ijms20030645