Map-Based Cloning and Functional Analysis of YE1 in Rice, Which Is Involved in Light-Dependent Chlorophyll Biogenesis and Photoperiodic Flowering Pathway
Abstract
:1. Introduction
2. Results
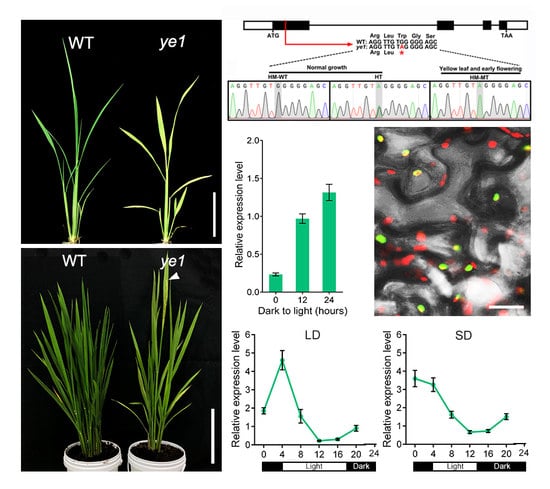
2.1. ye1 Mutant Exhibits Impaired Chloroplast Development and Photosynthesis
2.2. ye1 Mutant Displays Early Flowering
2.3. Map-Based Cloning of YE1 and Complementation Analysis
2.4. YE1 Protein Is Conserved in Land Plants
2.5. Expression Pattern and Subcellular Location of YE1
2.6. YE1 Is Involved in Light-Dependent Chl Synthesis
2.7. Diurnal Expression Patterns of Flowering Genes Are Altered in ye1
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Condition
4.2. Morphological, Physiological, and Cellular Analysis
4.3. Map-Based Cloning of YE1
4.4. Vector Construction and Transgenic Analysis
4.5. Expression Analysis
4.6. Subcellular Localization of YE1
4.7. Phylogenetic Analysis of YE1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fankhauser, C.; Chory, J. Light Control of Plant Development. Annu. Rev. Cell Dev. Biol. 1997, 13, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Kami, C.; Lorrain, S.; Hornitschek, P.; Fankhauser, C. Chapter Two-Light-Regulated Plant Growth and Development. In Current Topics in Developmental Biology; Timmermans, M.C.P., Ed.; Academic Press: Amsterdam, Netherland, 2010; Volume 91, pp. 29–66. [Google Scholar]
- Fromme, P.; Melkozernov, A.; Jordan, P.; Krauss, N. Structure and function of photosystem I: Interaction with its soluble electron carriers and external antenna systems. FEBS Lett. 2003, 555, 40–44. [Google Scholar] [CrossRef]
- Stenbaek, A.; Jensen, P.E. Redox regulation of chlorophyll biosynthesis. Phytochemistry 2010, 71, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Von Wettstein, D. Chlorophyll Biosynthesis. In Plant Molecular Biology 2; Herrmann, R.G., Larkins, B.A., Eds.; Springer: Boston, MA, USA, 1991; pp. 449–459. [Google Scholar] [Green Version]
- Suzuki, J.Y.; Bauer, C.E. A prokaryotic origin for light-dependent chlorophyll biosynthesis of plants. Proc. Natl. Acad. Sci. USA 1995, 92, 3749–3753. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, J.; Huang, L.; Leng, Y.; Dai, L.; Rao, Y.; Chen, L.; Wang, Y.; Tu, Z.; Hu, J.; et al. PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J. Exp. Bot. 2016, 67, 1297–1310. [Google Scholar] [CrossRef] [PubMed]
- Papenbrock, J.; Grimm, B. Regulatory network of tetrapyrrole biosynthesis-studies of intracellular signalling involved in metabolic and developmental control of plastids. Planta 2001, 213, 667–681. [Google Scholar] [CrossRef]
- Liu, D.; Kong, D.D.; Fu, X.K.; Ali, B.; Xu, L.; Zhou, W.J. Influence of exogenous 5-aminolevulinic acid on chlorophyll synthesis and related gene expression in oilseed rape de-etiolated cotyledons under water-deficit stress. Photosynthetica 2016, 54, 468–474. [Google Scholar] [CrossRef]
- Franklin, K.A.; Quail, P.H. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2010, 61, 11–24. [Google Scholar] [CrossRef]
- Casal, J.J. Phytochromes, Cryptochromes, Phototropin: Photoreceptor Interactions in Plants. Photochem. Photobiol. 2000, 71, 1–11. [Google Scholar] [CrossRef]
- McCormac, A.C.; Terry, M.J. Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J. 2002, 32, 549–559. [Google Scholar] [CrossRef]
- Moon, J.; Zhu, L.; Shen, H.; Huq, E. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 9433–9438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, C.; Müller, A.E. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009, 14, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lin, C. Light Regulation of Flowering Time in Arabidopsis. In Light Sensing in Plants; Wada, M., Shimazaki, K.-I., Iino, M., Eds.; Springer: Tokyo, Japan, 2005; pp. 325–332. [Google Scholar]
- Suárez-López, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Kardailsky, I.; Shukla, V.K.; Ahn, J.H.; Dagenais, N.; Christensen, S.K.; Nguyen, J.T.; Chory, J.; Harrison, M.J.; Weigel, D. Activation Tagging of the Floral Inducer FT. Science 1999, 286, 1962–1965. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Roussot, C.; Suárez-López, P.; Corbesier, L.; Vincent, C.; Piñeiro, M.; Hepworth, S.; Mouradov, A.; Justin, S.; Turnbull, C.; et al. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 2004, 131, 3615–3626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imaizumi, T.; Schultz, T.F.; Harmon, F.G.; Ho, L.A.; Kay, S.A. FKF1 F-Box Protein Mediates Cyclic Degradation of a Repressor of CONSTANS in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Hayama, R.; Coupland, G. The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 2004, 135, 677–684. [Google Scholar] [CrossRef]
- Shim, J.S.; Kubota, A.; Imaizumi, T. Circadian Clock and Photoperiodic Flowering in Arabidopsis: CONSTANS Is a Hub for Signal Integration. Plant Physiol. 2017, 173, 5–15. [Google Scholar] [CrossRef]
- Mockler, T.; Yang, H.; Yu, X.; Parikh, D.; Cheng, Y.-c.; Dolan, S.; Lin, C. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. USA 2003, 100, 2140–2145. [Google Scholar] [CrossRef] [Green Version]
- Valverde, F.; Mouradov, A.; Soppe, W.; Ravenscroft, D.; Samach, A.; Coupland, G. Photoreceptor Regulation of CONSTANS Protein in Photoperiodic Flowering. Science 2004, 303, 1003–1006. [Google Scholar] [CrossRef] [Green Version]
- Shekhawat, G.S.; Verma, K. Heme oxygenase (HO): An overlooked enzyme of plant metabolism and defence. J. Exp. Bot. 2010, 61, 2255–2270. [Google Scholar] [CrossRef] [PubMed]
- Terry, M.J.; Linley, P.J.; Kohchi, T. Making light of it: The role of plant heme oxygenases in phytochrome chromophore synthesis. Biochem. Soc. Transact. 2002, 30, 604–609. [Google Scholar] [CrossRef]
- Muramoto, T.; Kohchi, T.; Yokota, A.; Hwang, I.; Goodman, H.M. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 1999, 11, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Zheng, X.; Li, L.; Lin, W.; Xie, W.; Yang, J.; Chen, S.; Jin, W. Chlorophyll Deficiency in the Maize elongated mesocotyl2 Mutant Is Caused by a Defective Heme Oxygenase and Delaying Grana Stacking. PLoS ONE 2013, 8, e80107. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Cheng, Z.; Ma, X.; Wu, H.; Liu, Y.; Zhou, K.; Chen, Y.; Ma, W.; Bi, J.; Zhang, X.; et al. A knockdown mutation of YELLOW-GREEN LEAF2 blocks chlorophyll biosynthesis in rice. Plant Cell Rep. 2013, 32, 1855–1867. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Galbraith, D.W.; Talón, M.; Domingo, C. Analysis of PHOTOPERIOD SENSITIVITY5 Sheds Light on the Role of Phytochromes in Photoperiodic Flowering in Rice. Plant Physiol. 2009, 151, 681–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izawa, T.; Oikawa, T.; Tokutomi, S.; Okuno, K.; Shimamoto, K. Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J. 2000, 22, 391–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Gao, J.; Wan, C.; Zhang, F.; Xu, Z.; Huang, X.; Sun, X.; Deng, X. Divinyl Chlorophyll(ide) a Can Be Converted to Monovinyl Chlorophyll(ide) a by a Divinyl Reductase in Rice. Plant Physiol. 2010, 153, 994–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Wan, C.; Xu, Z.; Wang, P.; Wang, W.; Sun, C.; Ma, X.; Xiao, Y.; Zhu, J.; Gao, X.; et al. One Divinyl Reductase Reduces the 8-Vinyl Groups in Various Intermediates of Chlorophyll Biosynthesis in a Given Higher Plant Species, But the Isozyme Differs between Species. Plant Physiol. 2013, 161, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, J.-H.; Yoo, E.S.; Lee, C.-H.; Hirochika, H.; An, G. Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol. Biol. 2005, 57, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Ling, Y.; Fang, L.; Du, P.; Sang, X.; Zhao, F.; Li, Y.; Xie, R.; He, G. Gene cloning and functional analysis of yellow green leaf3 (ygl3) gene during the whole-plant growth stage in rice. Genes Genom. 2013, 35, 87–93. [Google Scholar] [CrossRef]
- Zhang, H.; Li, J.; Yoo, J.-H.; Yoo, S.-C.; Cho, S.-H.; Koh, H.-J.; Seo, H.S.; Paek, N.-C. Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol. Biol. 2006, 62, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.-J.; Zhang, H.-Q.; Wang, Y.; He, F.; Liu, J.-L.; Xiao, X.; Shu, Z.-F.; Li, W.; Wang, G.-H.; Wang, G.-L. Mapped Clone and Functional Analysis of Leaf-Color Gene Ygl7 in a Rice Hybrid (Oryza sativa L. ssp. indica). PLoS ONE 2014, 9, e99564. [Google Scholar] [CrossRef] [PubMed]
- Goh, C.-H.; Satoh, K.; Kikuchi, S.; Kim, S.-C.; Ko, S.-M.; Kang, H.-G.; Jeon, J.-S.; Kim, C.S.; Park, Y.-I. Mitochondrial activity in illuminated leaves of chlorophyll-deficient mutant rice (OsCHLH) seedlings. Plant Biotechnol. Rep. 2010, 4, 281–291. [Google Scholar] [CrossRef]
- Goh, C.H.; Oh, S.; An, G.; Moon, Y.H.; Lee, C.H. Activation of mitochondrial respiration in chlorophyll-deficient rice mutant seedlings. J. Plant Biol. 2007, 50, 430–439. [Google Scholar] [CrossRef]
- Ruan, B.; Gao, Z.; Zhao, J.; Zhang, B.; Zhang, A.; Hong, K.; Yang, S.; Jiang, H.; Liu, C.; Chen, G.; et al. The rice YGL gene encoding an Mg2+-chelatase ChlD subunit is affected by temperature for chlorophyll biosynthesis. J. Plant Biol. 2017, 60, 314–321. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Rahman, M.L.; Cho, S.-H.; Kim, Y.-S.; Koh, H.-J.; Yoo, S.-C.; Paek, N.-C. The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J. 2013, 74, 122–133. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Zhang, X.; He, B.; Diao, L.; Sheng, S.; Wang, J.; Guo, X.; Su, N.; Wang, L.; Jiang, L.; et al. A Chlorophyll-Deficient Rice Mutant with Impaired Chlorophyllide Esterification in Chlorophyll Biosynthesis. Plant Physiol. 2007, 145, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Jin, M.; Zheng, X.-M.; Chen, J.; Yuan, D.; Xin, Y.; Wang, M.; Huang, D.; Zhang, Z.; Zhou, K.; et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 16337–16342. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Xu, J.; Guo, H.; Jiang, L.; Chen, S.; Yu, C.; Zhou, Z.; Hu, P.; Zhai, H.; Wan, J. DTH8 Suppresses Flowering in Rice, Influencing Plant Height and Yield Potential Simultaneously. Plant Physiol. 2010, 153, 1747–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, H.; Ogiso-Tanaka, E.; Okumoto, Y.; Yoshitake, Y.; Izumi, H.; Yokoo, T.; Matsubara, K.; Hori, K.; Yano, M.; Inoue, H.; et al. Ef7 Encodes an ELF3-like Protein and Promotes Rice Flowering by Negatively Regulating the Floral Repressor Gene Ghd7 under Both Short- and Long-Day Conditions. Plant Cell Physiol. 2012, 53, 717–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; You, C.; Li, C.; Long, T.; Chen, G.; Byrne, M.E.; Zhang, Q. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 12915–12920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsubara, K.; Yamanouchi, U.; Wang, Z.-X.; Minobe, Y.; Izawa, T.; Yano, M. Ehd, a Rice Ortholog of the Maize INDETERMINATE Gene, Promotes Flowering by Upregulating Ehd1. Plant Physiol. 2008, 148, 1425–1435. [Google Scholar] [CrossRef]
- Takahashi, Y.; Teshima, K.M.; Yokoi, S.; Innan, H.; Shimamoto, K. Variations in Hd1 proteins, Hd3a promoters, and Ehd1expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl. Acad. Sci. USA 2009, 106, 4555. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2484. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Ogiso-Tanaka, E.; Matsubara, K.; Yamanouchi, U.; Ebana, K.; Yano, M. Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 2013, 76, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Hayama, R.; Yokoi, S.; Tamaki, S.; Yano, M.; Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422, 719. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Nonoue, Y.; Yano, M.; Izawa, T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 2010, 42, 635. [Google Scholar] [CrossRef]
- Izawa, T.; Mihara, M.; Suzuki, Y.; Gupta, M.; Itoh, H.; Nagano, A.J.; Motoyama, R.; Sawada, Y.; Yano, M.; Hirai, M.Y.; et al. Os-GIGANTEA Confers Robust Diurnal Rhythms on the Global Transcriptome of Rice in the Field. Plant Cell 2011, 23, 1741–1755. [Google Scholar] [CrossRef]
- Zhou, Y.; Gong, Z.; Yang, Z.; Yuan, Y.; Zhu, J.; Wang, M.; Yuan, F.; Wu, S.; Wang, Z.; Yi, C.; et al. Mutation of the Light-Induced Yellow Leaf 1 Gene, Which Encodes a Geranylgeranyl Reductase, Affects Chlorophyll Biosynthesis and Light Sensitivity in Rice. PLoS ONE 2013, 8, e75299. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Ren, Y.; Lv, J.; Wang, Y.; Liu, F.; Zhou, F.; Zhao, S.; Chen, S.; Peng, C.; Zhang, X.; et al. Young Leaf Chlorosis 1, a chloroplast-localized gene required for chlorophyll and lutein accumulation during early leaf development in rice. Planta 2013, 237, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Kim, E.-Y.; Han, S.-H.; Piao, W.; An, G.; Todaka, D.; Yamaguchi-Shinozaki, K.; Paek, N.-C. Rice Phytochrome-Interacting Factor-Like1 (OsPIL1) is involved in the promotion of chlorophyll biosynthesis through feed-forward regulatory loops. J. Exp. Bot. 2017, 68, 4103–4114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, C.-T.; Kim, S.-H.; Song, G.; Kim, D.; Paek, N.-C. Two NADPH: Protochlorophyllide Oxidoreductase (POR) Isoforms Play Distinct Roles in Environmental Adaptation in Rice. Rice 2017, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, J.; Rao, Y.; Zeng, Y.; Liu, H.; Zheng, T.; Zhang, G.; Hu, J.; Guo, L.; Qian, Q.; et al. Cloning and functional analysis of pale-green leaf (PGL10) in rice (Oryza sativa L.). Plant Growth Regul. 2016, 78, 69–77. [Google Scholar] [CrossRef]
- RYU, C.-H.; LEE, S.; CHO, L.-H.; KIM, S.L.; LEE, Y.-S.; CHOI, S.C.; JEONG, H.J.; YI, J.; PARK, S.J.; HAN, C.-D.; et al. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 2009, 32, 1412–1427. [Google Scholar] [CrossRef] [PubMed]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.L.; Lee, S.; Kim, H.J.; Nam, H.G.; An, G. OsMADS51 Is a Short-Day Flowering Promoter That Functions Upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 2007, 145, 1484–1494. [Google Scholar] [CrossRef] [Green Version]
- Komiya, R.; Ikegami, A.; Tamaki, S.; Yokoi, S.; Shimamoto, K. Hd3a and RFT1 are essential for flowering in rice. Development 2008, 135, 767–774. [Google Scholar] [CrossRef] [Green Version]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a Rice Ortholog of the Arabidopsis FT Gene, Promotes Transition to Flowering Downstream of Hd1 under Short-Day Conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Li, Q.; Xing, Y. Genes Contributing to Domestication of Rice Seed Traits and Its Global Expansion. Genes 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Kakizaki, T.; Matsumura, H.; Nakayama, K.; Che, F.-S.; Terauchi, R.; Inaba, T. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 2009, 151, 1339–1353. [Google Scholar] [CrossRef] [PubMed]
- Shimamoto, K.; Yokoi, S. The Photoperiodic Control of Flowering in Rice, a Short-Day Plant. In Light Sensing in Plants; Wada, M., Shimazaki, K.-I., Iino, M., Eds.; Springer: Tokyo, Japan, 2005; pp. 339–346. [Google Scholar]
- Goto, N.; Kumagai, T.; Koornneef, M. Flowering responses to light-breaks in photomorphogenic mutants of Arabidopsis thaliana, a long-day plant. Physiol. Plantar. 1991, 83, 209–215. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Su, Y.-S.; Lagarias, J.C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 2006, 57, 837–858. [Google Scholar] [CrossRef] [PubMed]
- Yanovsky, M.J.; Kay, S.A. Living by the calendar: How plants know when to flower. Nat. Rev. Mol. Cell Biol. 2003, 4, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Takano, M.; Inagaki, N.; Xie, X.; Yuzurihara, N.; Hihara, F.; Ishizuka, T.; Yano, M.; Nishimura, M.; Miyao, A.; Hirochika, H.; et al. Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 2005, 17, 3311–3325. [Google Scholar] [CrossRef] [PubMed]
- Bae, G.; Choi, G. Decoding of Light Signals by Plant Phytochromes and Their Interacting Proteins. Annu. Rev. Plant Biol. 2008, 59, 281–311. [Google Scholar] [CrossRef]
- Elich, T.D.; McDonagh, A.F.; Palma, L.A.; Lagarias, J.C. Phytochrome chromophore biosynthesis. Treatment of tetrapyrrole-deficient Avena explants with natural and non-natural bilatrienes leads to formation of spectrally active holoproteins. J. Biol. Chem. 1989, 264, 183–189. [Google Scholar]
- Chory, J.; Peto, C.A.; Ashbaugh, M.; Saganich, R.; Pratt, L.; Ausubel, F. Different Roles for Phytochrome in Etiolated and Green Plants Deduced from Characterization of Arabidopsis thaliana Mutants. Plant Cell 1989, 1, 867–880. [Google Scholar] [CrossRef] [Green Version]
- Weller, J.L.; Terry, M.J.; Rameau, C.; Reid, J.B.; Kendrick, R.E. The Phytochrome-Deficient pcd1 Mutant of Pea Is Unable to Convert Heme to Biliverdin IX[alpha]. The Plant cell 1996, 8, 55–67. [Google Scholar] [CrossRef] [Green Version]
- Cornah, J.E.; Terry, M.J.; Smith, A.G. Green or red: What stops the traffic in the tetrapyrrole pathway? Trends Plant Sci. 2003, 8, 224–230. [Google Scholar] [CrossRef]
- Kumar, A.M.; Söll, D. Antisense HEMA1 RNA expression inhibits heme and chlorophyll biosynthesis in arabidopsis. Plant Physiol. 2000, 122, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Pontoppidan, B.; Kannangara, C.G. Purification and partial characterisation of barley glutamyl-tRNAGlu reductase, the enzyme that directs glutamate to chlorophyll biosynthesis. Eur. J. Biochem. 1994, 225, 529–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terry, M.J.; Kendrick, R.E. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol. 1999, 119, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Goslings, D.; Meskauskiene, R.; Kim, C.; Lee, K.P.; Nater, M.; Apel, K. Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 2004, 40, 957–967. [Google Scholar] [CrossRef] [Green Version]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174. [Google Scholar] [CrossRef] [PubMed]
- Akhter, D.; Qin, R.; Nath, U.; Alamin, M.; Jin, X.; Shi, C. The Brown Midrib Leaf (bml) Mutation in Rice (Oryza sativa L.) Causes Premature Leaf Senescence and the Induction of Defense Responses. Genes 2018, 9, 203. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Xiao, Q.; Li, W.; Luo, T.; Yuan, G.; He, Z.; Liu, M.; Li, Q.; Xu, P.; Zhu, J.; et al. OsLAP6/OsPKS1, an orthologue of Arabidopsis PKSA/LAP6, is critical for proper pollen exine formation. Rice 2017, 10, 53. [Google Scholar] [CrossRef] [Green Version]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef]
- Dong, H.; Fei, G.-L.; Wu, C.-Y.; Wu, F.-Q.; Sun, Y.-Y.; Chen, M.-J.; Ren, Y.-L.; Zhou, K.-N.; Cheng, Z.-J.; Wang, J.-L.; et al. A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol. 2013, 162, 1867–1880. [Google Scholar] [CrossRef]
- Lichocka, M.; Schmelzer, E. Subcellular Localization Experiments and FRET-FLIM Measurements in Plants. Bioprotocol 2014, 4, e1018. [Google Scholar] [CrossRef]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, W695–W699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]









| Plant | ΦPSII | ETR | Fv/Fm | Pn (μmol·CO2·m−2·s−1) | Gs (mol·H2O·m−2·s−1) | Ci (μmol·CO2·m−2·s−1) | Tr (mmol·H2O·m−2·s−1) |
|---|---|---|---|---|---|---|---|
| WT | 0.74 ± 0.02 | 9.43 ± 0.05 | 0.84 ± 0.01 | 17.57 ± 0.21 | 0.90 ± 0.16 | 268.0 ± 6.55 | 10.62 ± 0.36 |
| yel | 0.71 ± 0.01 * | 6.93 ± 0.07 ** | 0.73 ± 0.01 ** | 5.65 ± 0.17 ** | 0.48 ± 0.09 ** | 264.0 ± 13.50 NS | 3.40 ± 0.22 ** |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Y.; Zou, T.; Li, L.; Tang, S.; Li, Q.; Zhang, J.; Chen, Y.; Wang, X.; Yang, G.; Hu, Y. Map-Based Cloning and Functional Analysis of YE1 in Rice, Which Is Involved in Light-Dependent Chlorophyll Biogenesis and Photoperiodic Flowering Pathway. Int. J. Mol. Sci. 2019, 20, 758. https://doi.org/10.3390/ijms20030758
Peng Y, Zou T, Li L, Tang S, Li Q, Zhang J, Chen Y, Wang X, Yang G, Hu Y. Map-Based Cloning and Functional Analysis of YE1 in Rice, Which Is Involved in Light-Dependent Chlorophyll Biogenesis and Photoperiodic Flowering Pathway. International Journal of Molecular Sciences. 2019; 20(3):758. https://doi.org/10.3390/ijms20030758
Chicago/Turabian StylePeng, Youlin, Ting Zou, Lamei Li, Shiwen Tang, Qiao Li, Jie Zhang, Yongjun Chen, Xuechun Wang, Guotao Yang, and Yungao Hu. 2019. "Map-Based Cloning and Functional Analysis of YE1 in Rice, Which Is Involved in Light-Dependent Chlorophyll Biogenesis and Photoperiodic Flowering Pathway" International Journal of Molecular Sciences 20, no. 3: 758. https://doi.org/10.3390/ijms20030758
APA StylePeng, Y., Zou, T., Li, L., Tang, S., Li, Q., Zhang, J., Chen, Y., Wang, X., Yang, G., & Hu, Y. (2019). Map-Based Cloning and Functional Analysis of YE1 in Rice, Which Is Involved in Light-Dependent Chlorophyll Biogenesis and Photoperiodic Flowering Pathway. International Journal of Molecular Sciences, 20(3), 758. https://doi.org/10.3390/ijms20030758