Characterization, Diversity, and Structure-Activity Relationship Study of Lipoamino Acids from Pantoea sp. and Synthetic Analogues
Abstract
:1. Introduction
2. Results and Discussion
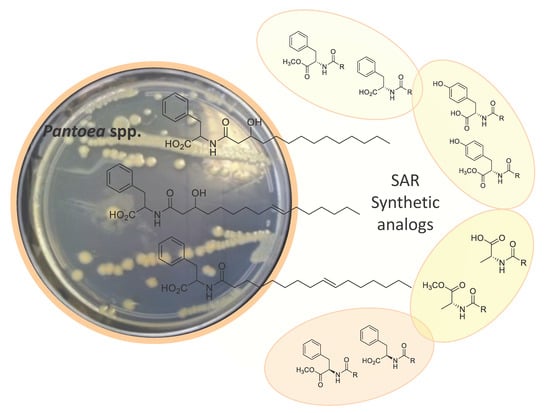
2.1. Isolation and Structure Elucidation
2.2. Synthesis of Lipoaminoacid Analogues
2.3. Biological Activities and SAR Investigation
2.4. Molecular Networking
3. Materials and Methods
3.1. General Procedures
3.2. Collection and Identification of Pantoea sp. SNB-VECD14B
3.3. Culture, Extraction, and Isolation
3.4. General Synthetic Procedure for Lipoamino Acid Methyl Ester Synthesis
3.5. General Synthetic Procedure for Lipoamino Acid Synthesis
3.6. Synthesis of Compound 3
3.7. Molecular Networking
3.7.1. Mass Spectrometry Analysis Parameters
3.7.2. MS Data Pretreatment
3.7.3. Molecular Networks Generation
3.8. Evaluation of Larvicidal Activity
3.8.1. Insect Collection and Rearing
3.8.2. Cup Assays
3.8.3. Tube Assays
3.8.4. Statistical Analysis
3.9. Antimicrobial Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACN | Acetonitrile |
| DMF | Dimethylformamide |
| DIEA | N,N-Diisopropylethylamine |
| EtOAc | Ethyl acetate |
| HATU | N-[(Dimethylamino)-1H-1,2,3-triazolo-[4,5-b]pyridin-1-ylmethylene]-N-methylmethanaminium hexafluorophosphate N-oxide |
| HCl | Hydrochloric acid |
| HPLC | High performance liquid chromatography |
| MH | Mueller hinton |
| MIC | Minimum inhibitory concentration |
| MN | Molecular networking |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| MS | Mass spectrometry |
| NMR | Nuclear magnetic resonance |
| PDA | Potato dextrose agar |
| RDBE | Round double bond equivalent |
| SAR | Structure activity relationship |
| UPLC | Ultra-high performance liquid chromatography |
| VRE | Vancomycin-resistant Enterococci |
| VRSA | Vancomycin-resistant Staphylococcus aureus |
| WHO | World Health Organization |
References
- Gould, I.M. The epidemiology of antibiotic resistance. Int. J. Antimicrob. Agents 2008, 32, S2–S9. [Google Scholar] [CrossRef] [PubMed]
- Pilacik, N.; Kaminska, T.; Augustynowicz-Kopec, E.; Krasowski, G. Etiology of bacterial infections and incidence of comorbidities in patients with tuberculosis, treated in Mazovian Treatment Centre of Tuberculosis and Lung Diseases during years 2012-2014. Wiad. Lek. 2015, 68, 347–353. [Google Scholar] [PubMed]
- Ventola, C.L. The Antibiotic Resistance Crisis. Part 1: Causes and Threats. Pharm. Ther 2015, 40, 277–283. [Google Scholar]
- Word Health Organization. Available online: http://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 18 September 2018).
- Kraemer, M.U.; Sinka, M.E.; Duda, K.A.; Mylne, A.Q.; Shearer, F.M.; Barker, C.M.; Moore, C.G.; Carvalho, R.G.; Coelho, G.E.; Van Bortel, W.; et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Ecol. Epidemiol. Glob. Health. 2015, 4, e08437. [Google Scholar] [CrossRef] [PubMed]
- Aktar, M.W.; Sengupta, D.; Chowdhury, A. Impact of pesticides use in agriculture: Their benefits and hazards. Interdiscip. Toxicol. 2009, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sokhna, C.; Ndiath, M.O.; Rogier, C. The changes in mosquito vector behaviour and the emerging resistance to insecticides will challenge the decline of malaria. Clin. Microbiol. Infect. 2013, 19, 902–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B. Entomopathogenic bacteria as a source of secondary metabolites. Curr. Opin. Chem. Biol. 2009, 13, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Frost, D.J.; Gunstone, F.D. The PMR analysis of non-conjugated alkenoic and alkynoic acids and esters. Chem. Phys. Lipids 1975, 15, 53–85. [Google Scholar] [CrossRef]
- Vast, A.; Singh, A.K.; Mukherjee, R.; Chopra, T.; Ravindran, M.S.; Mohanty, D.; Chatterji, D.; Reyrat, J.-M.; Gokhale, R.J. Retrobiosynthetic approach delineates the biosynthetic pathway and the structure of the acyl chain of mycobacterial glycopeptidolipids. J. Biol. Chem. 2012, 287, 30677–30687. [Google Scholar]
- Givaudin, S.A.; Staghouwer, H.; Thoen, C.; Van Buel, M. Improvements in or Relating to Organic Compounds, U.S. Patent 20160214928, 28 July 2016. [Google Scholar]
- Sheppard, G.S.; Wang, J.; Kawei, M.; BaMaung, N.Y.; Craig, R.A.; Erickson, S.A.; Lynch, L.; Patel, J.; Yang, F.; Searle, X.B.; et al. 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2. Biorg. Med. Chem. 2004, 14, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ©Cytoscape Consortium 2001-2017. Available online: https://cytoscape.org/ (accessed on 2 March 2018).
- World Health Organization. Guidelines for Laboratory and Field Testing of Mosquito Larvicides. Available online: http://apps.who.int/iris/bitstream/10665/69101/1/WHO_CDS_WHOPES_GCDPP_2005.13.pdf (accessed on 18 September 2018).
- Falkowski, M.; Jahn-Oyac, A.; Ferrero, E.; Issaly, J.; Eparvier, V.; Girod, R.; Rodrigues, A.M.; Stien, D.; Houël, E.; Dusfour, I. Assessment of A Simple Compound-Saving Method To Study Insecticidal Activity of Natural Extracts and Pure Compounds Against Mosquito Larvae. J. Am. Mosq. Control Assoc. 2016, 32, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Abott, W.S. A Method of Computing the Effectiveness of an Insecticide. J. Econ Entomol. 1925, 18, 265–267. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi, 2nd ed.; CLSI: Wayne, PA, USA, 2008. [Google Scholar]
- Clinical and Laboratory Standards. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 3rd ed.; CLSI: Wayne, PA, USA, 2008. [Google Scholar]
- Clinical and Laboratory Standards. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 8th ed.; CLSI: Wayne, PA, USA, 2009. [Google Scholar]
- Desbois, A.P.; Lebl, T.; Yan, L.; Smith, V.J. Isolation and structural characterisation of two antibacterial free fatty acids from the marine diatom, Phaeodactylum tricornutum. Appl. Microbiol. Biotechnol. 2008, 81, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Desbois, A.P.; Smith, V.J. Antibacterial free fatty acids: Activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 2010, 85, 1629–1642. [Google Scholar] [CrossRef] [PubMed]






| Compound 1 | Compound 2 | Compound 3 | ||||
|---|---|---|---|---|---|---|
| Position | δC a, type | δH b, m (J in Hz) | δC c, type | δH d, m (J in Hz) | δC a, type | δH b, m (J in Hz) |
| 1 | 176.0, C | 176.3, C | 176.5, C | |||
| 2 | 55.9, CH | 4.60, m | 57.6, CH | 4.53, m | 56.0, CH | 4.62, dd (8.0, 4.7) |
| 3 | 38.7, CH2 | 2.98, dd (13.9, 8.2) | 39.4, CH2 | 2.97, dd (14.0, 7.7) | 39.0, CH2 | 2.94, dd (13.8, 8.7) |
| 3.21, dd (13.9, 4.5) | 3.21, dd (14.0, 4.5) | 3.22, dd (13.8, 4.7) | ||||
| 4 | 139.0, C | 139.8, C | 139.2, C | |||
| 5/9 | 130.5, CH | 7.24, m | 130.7, CH | 7.24, m | 130.5, CH | 7.23, m |
| 6/8 | 129.3, CH | 7.25, m | 129.3, CH | 7.22, m | 129.4, CH | 7.24, m |
| 7 | 127.6, CH | 7.18, m | 127.3, CH | 7.15, m | 127.6, CH | 7.17, m |
| 1′ | 173.8, C | 173.5, C | 175.8, C | |||
| 2′ | 44.6, CH2 | 2.29, dd (14.4, 5.2) | 44.9, CH2 | 2.25, dd (14.4, 7.8) | 37.2, CH2 | 2.13, t (7.4) |
| 2.34, dd (14.4, 7.4) | 2.30, dd (14.5, 4.9) | 2.23, t (7.5) | ||||
| 3′ | 69.6, CH | 3.85, m | 67.9, CH | 3.85, m | 27.1, CH2 | 1.48, m |
| 4′ | 38.0, CH2 | 1.40, m | 38.2, CH2 | 1.39, m | 30.3, CH2 | 1.29, br s |
| 5′ | 30.9, CH2 | 1.29, br s | 31.0, CH2 | 1.30, m | 23.8, CH2 | 1.31, br s |
| 6′ | 30.9, CH2 | 1.29, br s | 31.0, CH2 | 1.30, m | 30.3, CH2 | 1.29, br s |
| 7′ | 30.9, CH2 | 1.29, br s | 31.0, CH2 | 1.30, m | 30.3, CH2 | 1.29, br s |
| 8′ | 30.9, CH2 | 1.29, br s | 28.3, CH2 | 2.04, m | 30.3, CH2 | 2.03, m |
| 9′ | 30.9, CH2 | 1.29, br s | 130.9, CH | 5.40, m | 131.0, CH | 5.35, m |
| 10′ | 30.9, CH2 | 1.29, br s | 130.9, CH | 5.40, m | 131.0, CH | 5.35, m |
| 11′ | 30.9, CH2 | 1.29, br s | 28.3, CH2 | 2.04, m | 30.3, CH2 | 2.03, m |
| 12′ | 30.9, CH2 | 1.29, br s | 31.0, CH2 | 1.30, m | 30.5, CH2 | 1.33, br s |
| 13′ | 23.6, CH2 | 1.31, m | 31.0, CH2 | 1.30, m | 30.5, CH2 | 1.33, br s |
| 14′ | 14.4, CH3 | 0.90, t (6.8) | 31.0, CH2 | 1.30, m | 30.5, CH2 | 1.33, br s |
| 15′ | 23.9, CH2 | 1.30, m | 23.8, CH2 | 1.31, br s | ||
| 16′ | 14.6, CH3 | 0.90, t (6.8) | 14.6, CH3 | 0.90, t (6.8) | ||
| Larvicidal Activity (% Mortality at 10 ppm) | MIC (μg/mL) | ||||
|---|---|---|---|---|---|
| Compound | C. albicans ATCC10231 | S. aureus ATCC29213 | MRSA ATCC33591 | T. rubrum SNB-TR1 | |
| 1 * | 32.1 | nd | − | 64 | nd |
| 2 * | nd | nd | − | 128 | nd |
| 3 */3 | 67.3/42.9 | nd/− | −/− | 16/64 | 128/− |
| ent-3 | 71.1 | nd | − | − | − |
| 3-OMe | 8.2 | − | − | − | − |
| ent-3-OMe | 20.4 | − | − | − | − |
| 4 | 0 | − | − | − | nd |
| 5 | 0 | >64 | − | >64 | nd |
| 6 | 2.2 | − | − | − | − |
| 7 | 10.6 | − | − | 8 | − |
| 8 | 0 | − | − | − | − |
| 9 | 10.2 | − | − | − | − |
| 10 | 6.2 | − | − | − | − |
| 11 | 6.0 | − | − | − | − |
| 12 | 2.1 | >64 | >64 | >64 | >256 |
| 13 | 4 | − | − | − | − |
| 14 | 1.9 | − | − | − | − |
| 15 | 0 | − | − | − | − |
| 16 | 1.8 | − | − | − | − |
| 17 | 5.5 | − | − | − | − |
| 18 | 3.8 | − | − | nd | 256 |
| 19 | 29.4 | − | − | − | 64 |
| 20 | 2.3 | − | − | − | − |
| 21 | 4.5 | − | − | − | − |
| 22 | 2.1 | − | 64 | 32 | 32 |
| 23 | 26.5 | − | − | − | 32 |
| 24 | 31.2 | − | − | − | − |
| 25 | 19.1 | − | − | − | − |
| 26 | 4.0 | − | − | − | − |
| 27 | 23.3 | − | − | − | nd |
| 28 | 4.5 | − | − | − | − |
| 29 | 6.1 | 64 | 64 | 64 | 32 |
| 30 | 18.7 | − | − | <8 | <8 |
| 31 | 53.3 | − | − | − | − |
| 32 | 49.0 | nd | − | − | − |
| 33 | 2.3 | − | − | − | − |
| 34 | 4.5 | nd | 32 | 32 | 32 |
| 35 | 4.5 | − | − | 32 | 32 |
| Pos. Ctrl.a | 40 | 4 | 0.2 | 0.6 | 4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Touré, S.; Desrat, S.; Pellissier, L.; Allard, P.-M.; Wolfender, J.-L.; Dusfour, I.; Stien, D.; Eparvier, V. Characterization, Diversity, and Structure-Activity Relationship Study of Lipoamino Acids from Pantoea sp. and Synthetic Analogues. Int. J. Mol. Sci. 2019, 20, 1083. https://doi.org/10.3390/ijms20051083
Touré S, Desrat S, Pellissier L, Allard P-M, Wolfender J-L, Dusfour I, Stien D, Eparvier V. Characterization, Diversity, and Structure-Activity Relationship Study of Lipoamino Acids from Pantoea sp. and Synthetic Analogues. International Journal of Molecular Sciences. 2019; 20(5):1083. https://doi.org/10.3390/ijms20051083
Chicago/Turabian StyleTouré, Seindé, Sandy Desrat, Léonie Pellissier, Pierre-Marie Allard, Jean-Luc Wolfender, Isabelle Dusfour, Didier Stien, and Véronique Eparvier. 2019. "Characterization, Diversity, and Structure-Activity Relationship Study of Lipoamino Acids from Pantoea sp. and Synthetic Analogues" International Journal of Molecular Sciences 20, no. 5: 1083. https://doi.org/10.3390/ijms20051083
APA StyleTouré, S., Desrat, S., Pellissier, L., Allard, P. -M., Wolfender, J. -L., Dusfour, I., Stien, D., & Eparvier, V. (2019). Characterization, Diversity, and Structure-Activity Relationship Study of Lipoamino Acids from Pantoea sp. and Synthetic Analogues. International Journal of Molecular Sciences, 20(5), 1083. https://doi.org/10.3390/ijms20051083