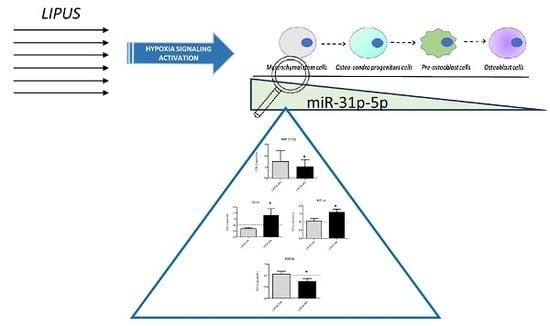
miR-31-5p Is a LIPUS-Mechanosensitive MicroRNA that Targets HIF-1α Signaling and Cytoskeletal Proteins
Abstract
:1. Introduction
2. Results
2.1. LIPUS Stimulation Induces the Modulation of Cytoskeletal Proteins
2.2. LIPUS Stimulation Induces the Expression of HIF-1α
2.3. LIPUS Stimulation Promotes MiR-31-5p Expression
2.4. The Presence of the miR-31-5p Is Useful to Induce LIPUS Effects on hMSCs
2.5. The Role of miR-31-5p on hMSCs Osteoblast Differentiation
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. LIPUS Treatment
4.3. Cell Transfection
4.4. hMSC Viability (WST-1 Test)
4.5. RNA Extraction and Real-Time PCR
4.6. ELISA Assay
4.7. TransAM Kit
4.8. Western Blot Analysis
4.9. Statistical Analysis
4.10. Data Availability
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Azuma, Y.; Ito, M.; Harada, Y.; Takagi, H.; Ohta, T.; Jingushi, S. Low-intensity pulsed ultrasound accelerates rat femoral fracture healing by acting on the various cellular reactions in the fracture callus. J. Bone Miner. Res. 2001, 16, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Yang, X.; Wei, X.; Chen, J.; Fu, N.; Fu, Y.; Ba, K.; Li, G.; Yao, Y.; Liang, C.; et al. Osteogenic differentiation of adipose-derived stem cells prompted by low-intensity pulsed ultrasound. Cell Prolif. 2013, 46, 320–327. [Google Scholar] [CrossRef]
- Costa, V.; Carina, V.; Fontana, S.; De Luca, A.; Monteleone, F.; Pagani, S.; Sartori, M.; Setti, S.; Faldini, C.; Alessandro, R.; et al. Osteogenic commitment and differentiation of human mesenchymal stem cells by low-intensity pulsed ultrasound stimulation. J. Cell. Physiol. 2018, 233, 1558–1573. [Google Scholar] [CrossRef]
- Fang, W.B.; Ireton, R.C.; Zhuang, G.; Takahashi, T.; Reynolds, A.; Chen, J. Overexpression of EPHA2 receptor destabilizes adherens junctions via a RhoA-dependent mechanism. J. Cell Sci. 2008, 121, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Carina, V.; Costa, V.; Raimondi, L.; Pagani, S.; Sartori, M.; Figallo, E.; Setti, S.; Alessandro, R.; Fini, M.; Giavaresi, G. Effect of low-intensity pulsed ultrasound on osteogenic human mesenchymal stem cells commitment in a new bone scaffold. J. Appl. Biomater. Funct. Mater. 2017, 15, e215–e222. [Google Scholar] [CrossRef]
- Kusuyama, J.; Seong, C.H.; Bandow, K.; Kakimoto, K.; Ohnishi, T.; Matsuguchi, T. Low intensity pulsed ultrasound (LIPUS) helps to maintain the undifferentiated status of mesenchymal stem cells. J. Orthop. Trauma 2015, 29, S2. [Google Scholar] [CrossRef]
- Pacary, E.; Tixier, E.; Coulet, F.; Roussel, S.; Petit, E.; Bernaudin, M. Crosstalk between HIF-1 and ROCK pathways in neuronal differentiation of mesenchymal stem cells, neurospheres and in PC12 neurite outgrowth. Mol. Cell. Neurosci. 2007, 35, 409–423. [Google Scholar] [CrossRef]
- Raheja, L.F.; Genetos, D.C.; Wong, A.; Yellowley, C.E. Hypoxic regulation of mesenchymal stem cell migration: The role of RhoA and HIF-1α. Cell Biol. Int. 2011, 35, 981–989. [Google Scholar] [CrossRef]
- Wheeler, A.P.; Ridley, A.J. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp. Cell Res. 2004, 301, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Vertelov, G.; Kharazi, L.; Muralidhar, M.G.; Sanati, G.; Tankovich, T.; Kharazi, A. High targeted migration of human mesenchymal stem cells grown in hypoxia is associated with enhanced activation of RhoA. Stem Cell Res. Ther. 2013, 4, 5. [Google Scholar] [CrossRef]
- Güntert, T.; Gassmann, M.; Ogunshola, O.O. Temporal Rac1—HIF-1 crosstalk modulates hypoxic survival of aged neurons. Brain Res. 2016, 1642, 298–307. [Google Scholar]
- Yin, C.P.; Guan, S.H.; Zhang, B.; Wang, X.X.; Yue, S.W. Upregulation of HIF-1α protects neuroblastoma cells from hypoxia-induced apoptosis in a RhoA-dependent manner. Mol. Med. Rep. 2015, 12, 7123–7131. [Google Scholar] [CrossRef]
- Dispenza, C.; Sabatino, M.; Ajovalasit, A.; Ditta, L.; Ragusa, M.; Purrello, M.; Costa, V.; Conigliaro, A.; Alessandro, R. Nanogel-antimiR-31 conjugates affect colon cancer cells behaviour. RSC Adv. 2017, 7, 52039–52047. [Google Scholar] [CrossRef]
- Turcotte, S.; Desrosiers, R.R.; Béliveau, R. HIF-1alpha mRNA and protein upregulation involves Rho GTPase expression during hypoxia in renal cell carcinoma. J. Cell Sci. 2003, 116, 2247–2260. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Raimondi, L.; Conigliaro, A.; Salamanna, F.; Carina, V.; De Luca, A.; Bellavia, D.; Alessandro, R.; Fini, M.; Giavaresi, G. Hypoxia-inducible factor 1A may regulate the commitment of mesenchymal stromal cells toward angio-osteogenesis by mirna-675-5P. Cytotherapy 2017, 19, 1412–1425. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Gao, D.; Gao, C.; Wei, P.; Niu, M.; Shuai, C. MicroRNAs regulate signaling pathways in osteogenic differentiation of mesenchymal stem cells (Review). Mol. Med. Rep. 2016, 14, 623–629. [Google Scholar] [CrossRef]
- Wang, J.; Wang, C.D.; Zhang, N.; Tong, W.X.; Zhang, Y.F.; Shan, S.Z.; Zhang, X.L.; Li, Q.F. Mechanical stimulation orchestrates the osteogenic differentiation of human bone marrow stromal cells by regulating HDAC1. Cell Death Dis. 2016, 7, e2221. [Google Scholar] [CrossRef]
- Chen, T.; Yao, L.Q.; Shi, Q.; Ren, Z.; Ye, L.C.; Xu, J.M.; Zhou, P.H.; Zhong, Y.S. MicroRNA-31 contributes to colorectal cancer development by targeting factor inhibiting HIF-1α (FIH-1). Cancer Biol. Ther. 2014, 15, 516–523. [Google Scholar] [CrossRef]
- Lo Dico, A.; Costa, V.; Martelli, C.; Diceglie, C.; Rajata, F.; Rizzo, A.; Mancone, C.; Tripodi, M.; Ottobrini, L.; Alessandro, R.; et al. MiR675-5p acts on HIF-1α to sustain hypoxic responses: A new therapeutic strategy for glioma. Theranostics 2016, 6, 1105–1118. [Google Scholar] [CrossRef]
- Xie, Q.; Wang, Z.; Bi, X.; Zhou, H.; Wang, Y.; Gu, P.; Fan, X. Effects of miR-31 on the osteogenesis of human mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2014, 446, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Baglìo, S.R.; Devescovi, V.; Granchi, D.; Baldini, N. MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene 2013, 527, 321–331. [Google Scholar] [CrossRef]
- Bellavia, D.; De Luca, A.; Carina, V.; Costa, V.; Raimondi, L.; Salamanna, F.; Alessandro, R.; Fini, M.; Giavaresi, G. Deregulated miRNAs in bone health: Epigenetic roles in osteoporosis. Bone 2019. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Bi, X.; Zhou, H.; You, Z.; Wang, Y.; Gu, P.; Fan, X. Repair of critical-sized bone defects with anti-miR-31-expressing bone marrow stromal stem cells and poly(glycerol sebacate) scaffolds. Eur. Cells Mater. 2014, 27, 13–24. [Google Scholar] [CrossRef]
- Deng, Y.; Wu, S.; Zhou, H.; Bi, X.; Wang, Y.; Hu, Y.; Gu, P.; Fan, X. Effects of a miR-31, Runx2, and Satb2 regulatory loop on the osteogenic differentiation of bone mesenchymal stem cells. Stem Cells Dev. 2013, 22, 2278–2286. [Google Scholar] [CrossRef] [PubMed]
- Manochantr, S.; Marupanthorn, K.; Tantrawatpan, C.; Kheolamai, P.; Tantikanlayaporn, D.; Sanguanjit, P. The effects of BMP-2, miR-31, miR-106a, and miR-148a on osteogenic differentiation of MSCs derived from amnion in comparison with MSCs derived from the bone marrow. Stem Cells Int. 2017, 2017, 7257628. [Google Scholar] [CrossRef]
- Wang, H.; Sun, Z.; Wang, Y.; Hu, Z.; Zhou, H.; Zhang, L.; Hong, B.; Zhang, S.; Cao, X. miR-33-5p, a novel mechano-sensitive microRNA promotes osteoblast differentiation by targeting Hmga2. Sci. Rep. 2016, 6, 23170. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- McCully, M.; Conde, J.; Baptista, P.; Mullin, M.; Dalby, M.J.; Berry, C.C. Nanoparticle-antagomiR based targeting of miR-31 to induce osterix and osteocalcin expression in mesenchymal stem cells. PLoS ONE 2018, 13, e0192562. [Google Scholar] [CrossRef]
- Erdogan, O.; Esen, E. Biological aspects and clinical importance of ultrasound therapy in bone healing. J. Ultrasound Med. 2009, 28, 765–776. [Google Scholar] [CrossRef]
- Zieseniss, A. Hypoxia and the modulation of the actin cytoskeleton—Emerging interrelations. Hypoxia 2014, 2, 11–21. [Google Scholar] [CrossRef]
- Gilkes, D.M.; Xiang, L.; Lee, S.J.; Chaturvedi, P.; Hubbi, M.E.; Wirtz, D.; Semenza, G.L. Hypoxia-inducible factors mediate coordinated RhoA-ROCK1 expression and signaling in breast cancer cells. Proc. Natl. Acad. Sci. USA 2014, 111, E384–E393. [Google Scholar] [CrossRef]
- Xue, Y.; Bi, F.; Zhang, X.; Zhang, S.; Pan, Y.; Liu, N.; Shi, Y.; Yao, X.; Zheng, Y.; Fan, D. Role of Rac1 and Cdc42 in hypoxia induced p53 and von Hippel-Lindau suppression and HIF1alpha activation. Int. J. Cancer 2006, 118, 2965–2972. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Semenza, G.L. Rac1 activity is required for the activation of hypoxia-inducible factor 1. J. Biol. Chem. 2001, 276, 21166–21172. [Google Scholar] [CrossRef]
- Zhang, X.B.; Song, L.; Wen, H.J.; Bai, X.X.; Li, Z.J.; Ma, L.J. Upregulation of microRNA-31 targeting integrin α5 suppresses tumor cell invasion and metastasis by indirectly regulating PI3K/AKT pathway in human gastric cancer SGC7901 cells. Tumour. Biol. 2016, 37, 8317–8325. [Google Scholar] [CrossRef]
- Stepicheva, N.A.; Song, J.L. Function and regulation of microRNA-31 in development and disease. Mol. Reprod. Dev. 2016, 83, 654–674. [Google Scholar] [CrossRef] [PubMed]
- Dobreva, G.; Chahrour, M.; Dautzenberg, M.; Chirivella, L.; Kanzler, B.; Farinas, I.; Karsenty, G.; Grosschedl, R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 2006, 125, 971–986. [Google Scholar] [CrossRef]
- Costa, V.; Lo Dico, A.; Rizzo, A.; Rajata, F.; Tripodi, M.; Alessandro, R.; Conigliaro, A. MiR-675-5p supports hypoxia induced epithelial to mesenchymal transition in colon cancer cells. Oncotarget 2017, 8, 24292–24302. [Google Scholar] [CrossRef]
- Yang, C.; Huntoon, K.; Ksendzovsky, A.; Zhuang, Z.; Lonser, R.R. Proteostasis modulators prolong missense VHL protein activity and halt tumor progression. Cell Rep. 2013, 3, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Masoud, G.N.; Li, W. HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 2015, 5, 378–389. [Google Scholar] [CrossRef]
- Wang, Y.; Wan, C.; Deng, L.; Liu, X.; Cao, X.; Gilbert, S.R.; Bouxsein, M.L.; Faugere, M.C.; Guldberg, R.E.; Gerstenfeld, L.C.; et al. The hypoxia-inducible factor alpha pathway couples angiogenesis to osteogenesis during skeletal development. J. Clin. Investig. 2007, 117, 1616–1626. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- R Foundation for Statistical Computing. A Language and Environment for Statistical Computing. 2008. Available online: http://www.R-project.org (accessed on 28 March 2019).







| Gene | Primer Forward | Primer Reverse |
|---|---|---|
| HIF-1A “Hypoxia-inducible factor 1-alpha” | TGATTGCATCTCCATCTCCTACC | GACTCAAAGCGACAGATAACACG |
| HIF-1AN “Hypoxia-inducible factor 1-alpha inhibitor” | TGGGGGCAGCTTACCTCTAA | TGGGTAGAGGCACTCGAAC |
| RAC-1 “Ras-related C3 botulinum toxin substrate 1” | TGAAAGCCTTCAGTCCCGTG | TGGTGATGCAGGCTGAACAAT |
| RHOA “Transforming protein RhoA” | GAAAACCGGTGAATCTGCGC | AGAACACATCTGTTTGCGGA |
| VEGF “Vascular endothelial growth factor” | CGAGGGCCTGGAGTGTGT | CGCATAATCTGCATGGTGATG |
| VHL “Von Hippel-Lindau disease tumor suppressor” | GACGGACAGCCTATTTTTGCC | TCCCATCCGTTGATGTGCAA |
| SOX9 “Transcription factor SOX-9” | GACTTCTGAACGAGAGCGAGA | CGTTCTTCACCGACTTCCTC |
| Reference Gene | ||
| ACTB “Beta-actin” | ATCAAGATCATTGCTCCTCCTGA | CTGCTTGCTGATCCACATCTG |
| Gene | Qiagen Primers | Catalog Number |
|---|---|---|
| RUNX2 | Hs_RUNX2_1_SG-QuantiTect Primer Assay | QT00020517 |
| ALPL | Hs_ALPL_1_SG-QuantiTect Primer Assay | QT00012957 |
| BGLAP | Hs_BGLAP_1_SG-QuantiTect Primer Assay | QT00232771 |
| SPP1 | Hs_SPP1_1_SG-QuantiTect Primer Assay | QT01008798 |
| SP7 | Hs-SP7_1_SG-QuantiTect Primer Assay | QT00213514 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, V.; Carina, V.; Conigliaro, A.; Raimondi, L.; De Luca, A.; Bellavia, D.; Salamanna, F.; Setti, S.; Alessandro, R.; Fini, M.; et al. miR-31-5p Is a LIPUS-Mechanosensitive MicroRNA that Targets HIF-1α Signaling and Cytoskeletal Proteins. Int. J. Mol. Sci. 2019, 20, 1569. https://doi.org/10.3390/ijms20071569
Costa V, Carina V, Conigliaro A, Raimondi L, De Luca A, Bellavia D, Salamanna F, Setti S, Alessandro R, Fini M, et al. miR-31-5p Is a LIPUS-Mechanosensitive MicroRNA that Targets HIF-1α Signaling and Cytoskeletal Proteins. International Journal of Molecular Sciences. 2019; 20(7):1569. https://doi.org/10.3390/ijms20071569
Chicago/Turabian StyleCosta, Viviana, Valeria Carina, Alice Conigliaro, Lavinia Raimondi, Angela De Luca, Daniele Bellavia, Francesca Salamanna, Stefania Setti, Riccardo Alessandro, Milena Fini, and et al. 2019. "miR-31-5p Is a LIPUS-Mechanosensitive MicroRNA that Targets HIF-1α Signaling and Cytoskeletal Proteins" International Journal of Molecular Sciences 20, no. 7: 1569. https://doi.org/10.3390/ijms20071569
APA StyleCosta, V., Carina, V., Conigliaro, A., Raimondi, L., De Luca, A., Bellavia, D., Salamanna, F., Setti, S., Alessandro, R., Fini, M., & Giavaresi, G. (2019). miR-31-5p Is a LIPUS-Mechanosensitive MicroRNA that Targets HIF-1α Signaling and Cytoskeletal Proteins. International Journal of Molecular Sciences, 20(7), 1569. https://doi.org/10.3390/ijms20071569