A Small In Vitro Fermentation Model for Screening the Gut Microbiota Effects of Different Fiber Preparations
Abstract
:1. Introduction
2. Results
2.1. Fiber Characteristics
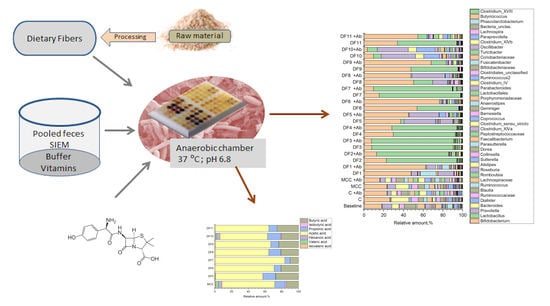
2.2. Fecal Fermentations of Commercial Dietary Fibers
2.3. Fecal Fermentations of Experimental Dietary Fiber Preparations
2.4. Fecal Fermentations of Dietary Fiber Preparations in the Presence of Amoxicillin-Clavulanate
3. Discussion
4. Materials and Methods
4.1. Fibers and Their Characterization
4.2. The In Vitro Fermentation Static Model
4.2.1. Preparation of Fecal Inoculum
4.2.2. Experimental Setup
4.3. Analyses of Fecal Samples
4.3.1. Short Chain Fatty Acid Analysis
4.3.2. DNA Extraction
4.3.3. Bacterial Quantification Using Real-Time qPCR (qPCR)
4.3.4. 16S rRNA Gene Amplicon Sequencing
4.3.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Turck, D.; Bernet, J.-P.; Marx, J.; Kempf, H.; Giard, P.; Walbaum, O.; Lacombe, A.; Rembert, F.; Toursel, F.; Bernasconi, P.; et al. Incidence and risk factors of oral antibiotic-associated diarrhea in an outpatient pediatric population. J. Pediatr. Gastroenterol. Nutr. 2003, 37, 22–26. [Google Scholar] [CrossRef]
- McFarland, L.V. Antibiotic-associated diarrhea: Epidemiology, trends and treatment. Future Microbiol. 2008, 3, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Chai, G.; Governale, L.; McMahon, A.W.; Trinidad, J.P.; Staffa, J.; Murphy, D. Trends of outpatient prescription drug utilization in US children, 2002–2010. Pediatrics 2012, 130, 23–31. [Google Scholar] [CrossRef]
- Korpela, K.; Salonen, A.; Virta, L.J.; Kekkonen, R.A.; Forslund, K.; Bork, P.; De Vos, W.M. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 2016, 7, 10410. [Google Scholar] [CrossRef] [PubMed]
- Lange, K.; Buerger, M.; Stallmach, A.; Bruns, T. Effects of antibiotics on gut microbiota. Dig. Dis. 2016, 34, 260–268. [Google Scholar] [CrossRef]
- Conlon, M.; Bird, A. The impact of diet and lifestyle on gut microbiota and human health. Nutrients 2015, 7, 17–44. [Google Scholar] [CrossRef]
- Holscher, H.D. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 2017, 8, 172–184. [Google Scholar] [CrossRef]
- Nordlund, E.; Aura, A.M.; Mattila, I.; Kössö, T.; Rouau, X.; Poutanen, K. Formation of phenolic microbial metabolites and short-chain fatty acids from rye, wheat, and oat bran and their fractions in the metabolical in vitro colon model. J. Agric. Food Chem. 2012, 60, 8134–8145. [Google Scholar] [CrossRef]
- Korpela, K. Diet, microbiota, and metabolic health: Trade-off between saccharolytic and proteolytic fermentation. Annu. Rev. Food Sci. Technol. 2018, 9, 65–84. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Lobel, L.; Garrett, W.S. Butyrate makes macrophages “Go Nuclear” against bacterial pathogens. Immunity 2019, 50, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Geirnaert, A.; Calatayud, M.; Grootaert, C.; Laukens, D.; Devriese, S.; Smagghe, G.; De Vos, M.; Boon, N.; Van de Wiele, T. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 2017, 7, 11450. [Google Scholar] [CrossRef]
- Gill, P.A.; van Zelm, M.C.; Muir, J.G.; Gibson, P.R. Short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment. Pharm. Ther. 2018, 48, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Redondo-Blanco, S.; Gutierrez-del-Rio, I.; Miguélez, E.M.; Villar, C.J.; Lombo, F. Colon microbiota fermentation of dietary prebiotics towards short-chain fatty acids and their roles as anti-inflammatory and antitumour agents: A review. J. Funct. Foods 2016, 25, 511–522. [Google Scholar] [CrossRef]
- Míguez, B.; Gómez, B.; Parajó, J.C.; Alonso, J.L. Potential of fructooligosaccharides and xylooligosaccharides as substrates to counteract the undesirable effects of several antibiotics on elder fecal microbiota: A first in vitro approach. J. Agric. Food Chem. 2018, 66, 9426–9437. [Google Scholar] [CrossRef]
- Soldi, S.; Vasileiadis, S.; Lohner, S.; Uggeri, F.; Puglisi, E.; Molinari, P.; Donner, E.; Sieland, C.; Decsi, T.; Sailer, M.; et al. Prebiotic supplementation over a cold season and during antibiotic treatment specifically modulates the gut microbiota composition of 3–6 year-old children. Benef. Microbes 2019, 1–12. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S. Models for intestinal fermentation: Association between food components, delivery systems, bioavailability and functional interactions in the gut. Curr. Opin. Biotechnol. 2007, 18, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.N.; Zihler, A.; Chassard, C.; Lacroix, C. Advances and perspectives in in vitro human gut fermentation modeling. Trends Biotechnol. 2012, 30, 17–25. [Google Scholar] [CrossRef]
- Venema, K.; van den Abbeele, P. Experimental models of the gut microbiome. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 115–126. [Google Scholar] [CrossRef]
- Muttakin, S.; Moxon, T.E.; Gouseti, O. In vivo, in vitro, and in silico studies of the GI tract. In Interdisciplinary Approaches to Food Digestion; Gouseti, O., Bornhorst, G.M., Bakalis, S., Mackie, A., Eds.; Springer: Cham, Switzerland, 2019; pp. 29–67. [Google Scholar]
- Elzinga, J.; van der Oost, J.; de Vos, W.M.; Smidt, H. The use of defined microbial communities to model host-microbe interactions in the human gut. Microbiol. Mol. Biol. Rev. 2019, 83, e00054-18. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Molly, K.; Van de Woestyne, M.; Verstraete, W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 1993, 39, 254–258. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Gibson, G.R.; Cummings, J.H. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 1992, 72, 57–64. [Google Scholar] [PubMed]
- Macfarlane, G.T.; Macfarlane, S.; Gibson, G.R. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb. Ecol. 1998, 35, 180–187. [Google Scholar] [CrossRef]
- Minekus, M.; Smeets-Peeters, M.F.; Bernalier, A.F.; Marol-Bonnin, S.F.; Havenaar, R.F.; Marteau, P.F.; Alric, M.F.; Fonty, G.; Huis in’t Veld, J.H.J. A computer-controlled system to simulate conditions of the large intestine with peristaltic mixing, water absorption and absorption of fermentation products. Appl. Microbiol. Biotechnol. 1999, 53, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Fehlbaum, S.; Chassard, C.; Haug, M.C.; Fourmestraux, C.; Derrien, M.; Lacroix, C. Design and investigation of PolyFermS in vitro continuous fermentation models inoculated with immobilized fecal microbiota mimicking the elderly colon. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.F.; Walton, G.E.; Jiang, L.; Plummer, S.; Garaiova, I.; Gibson, G.R. Comparative analysis of intestinal tract models. Annu. Rev. Food. Sci. Technol. 2015, 6, 329–350. [Google Scholar] [CrossRef]
- Wiese, M.; Khakimov, B.; Nielsen, S.; Sørensen, H.; van den Berg, F.; Nielsen, D.S. CoMiniGut—A small volume in vitro colon model for the screening of gut microbial fermentation processes. PeerJ 2018, 6, e4268. [Google Scholar] [CrossRef]
- ODonnell, M.; Rea, M.C.; Shanahan, F.; Ross, P. The use of a mini-bioreactor fermentation system as a reproducible, high-throughput ex-vivo batch model of the distal colon. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Aura, A.M.; Maukonen, J. One Compartment Fermentation Model. In The Impact of Food Bioactives on Health; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, S., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham, Switzerland, 2015; pp. 281–292. [Google Scholar]
- Barry, J.L.; Hoebler, C.; Macfarlane, G.T.; Macfarlane, S.; Mathers, J.C.; Reed, K.A.; Mortensen, P.B.; Nordgaard, I.; Rowland, I.R.; Rumney, C.J. Estimation of the fermentability of dietary fiber in vitro: A European interlaboratory study. Br. J. Nutr. 1995, 74, 303–322. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, M.; Ramiro-Garcia, J.; Koenen, M.E.; Venema, K. To pool or not to pool? Impact of the use of individual and pooled fecal samples for in vitro fermentation studies. J. Microbiol. Methods 2014, 107, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vamanu, E.; Gatea, F.; Sârbu, I. In vitro ecological response of the human gut microbiome to bioactive extracts from edible wild mushrooms. Molecules 2018, 23, 2128. [Google Scholar] [CrossRef] [PubMed]
- Martínez, I.; Kim, J.; Duffy, P.R.; Schlegel, V.L.; Walter, J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS ONE 2010, 5, e15046. [Google Scholar] [CrossRef]
- Venkataraman, A.; Sieber, J.R.; Schmidt, A.W.; Waldron, C.; Theis, K.R.; Schmidt, T.M. Variable responses of human microbiomes to dietary supplementation with resistant starch. Microbiome 2016, 4, 33. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.V.; Lucio, M.; Lee, L.H.; VerBerkmoes, N.C.; Brislawn, C.J.; Bernhardt, J.; Lamendella, R.; McDermott, J.E.; Bergeron, N.; Heinzmann, S.S.; et al. Impact of dietary resistant starch on the human gut microbiome, metaproteome, and metabolome. mBio 2017, 8, e01343-17. [Google Scholar] [CrossRef] [PubMed]
- Vital, M.; Howe, A.; Bergeron, N.; Krauss, R.M.; Jansson, J.K.; Tiedje, J.M. Metagenomic insights into the degradation of resistant starch by human gut microbiota. Appl. Environ. Microbiol. 2018, 84, e01562-18. [Google Scholar] [CrossRef]
- Ramirez-Farias, C.; Slezak, K.; Fuller, Z.; Duncan, A.; Holtrop, G.; Louis, P. Effect of inulin on the human gut microbiota: Stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 2008, 101, 541–550. [Google Scholar] [CrossRef]
- Costabile, A.; Kolida, S.; Klinder, A.; Gietl, E.; Bäuerlein, M.; Frohberg, C.; Landschütze, V.; Gibson, G.R. A double-blind, placebo-controlled, cross-over study to establish the bifidogenic effect of a very-long-chain inulin extracted from globe artichoke (Cynara scolymus) in healthy human subjects. Br. J. Nutr. 2010, 104, 1007–1017. [Google Scholar] [CrossRef]
- Dewulf, E.M.; Cani, P.D.; Claus, S.P.; Fuentes, S.; Puylaert, P.G.; Neyrinck, A.M.; Possemiers, S.; Van Holle, A.; Muccioli, G.G.; Deldicque, L.; et al. Insight into the prebiotic concept: Lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 2013, 62, 1112–1121. [Google Scholar] [CrossRef]
- Healey, G.; Murphy, R.; Butts, C.; Brough, L.; Whelan, K.; Coad, J. Habitual dietary fibre intake influences gut microbiota response to an inulin-type fructan prebiotic: A randomised, double-blind, placebo-controlled, cross-over, human intervention study. Br. J. Nutr. 2018, 119, 176–189. [Google Scholar] [CrossRef]
- Fehlbaum, S.; Prudence, K.; Kieboom, J.; Heerikhuisen, M.; van den Broek, T.; Schuren, F.; Steinert, R.E.; Raederstorff, D. In vitro fermentation of selected prebiotics and their effects on the composition and activity of the adult gut microbiota. Int. J. Mol. Sci. 2018, 19, 3097. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Corradini, C.; Amaretti, A.; Nicolini, M.; Pompei, A.; Zanoni, S.; Matteuzzi, D. Fermentation of fructooligosaccharides and inulin by bifidobacteria: A comparative study of pure and fecal cultures. Appl. Environ. Microbiol. 2005, 71, 6150–6158. [Google Scholar] [CrossRef]
- Van De Wiele, T.; Boon, N.; Possemiers, S.; Jacobs, H.; Verstraete, W. Inulin-type fructans of longer degree of polymerization exert more pronounced in vitro prebiotic effects. J. Appl. Microbiol. 2007, 102, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Selak, M.; Rivière, A.; Moens, F.; Van den Abbeele, P.; Geirnaert, A.; Rogelj, I.; Leroy, F.; De Vuyst, L. Inulin-type fructan fermentation by bifidobacteria depends on the strain rather than the species and region in the human intestine. Appl. Microbiol. Biotechnol. 2016, 100, 4097–4107. [Google Scholar] [CrossRef]
- Pompei, A.; Cordisco, L.; Raimondi, S.; Amaretti, A.; Pagnoni, U.M.; Matteuzzi, D.; Rossi, M. In vitro comparison of the prebiotic effects of two inulin-type fructans. Anaerobe 2008, 14, 280–286. [Google Scholar] [CrossRef]
- Figueroa-Espinoza, M.C.; Poulsen, C.; Borch Søe, J.; Zargahi, M.R.; Rouau, X. Enzymatic solubilization of arabinoxylans from native, extruded, and high-shear-treated rye bran by different endo-xylanases and other hydrolyzing enzymes. J. Agric. Food Chem. 2004, 52, 4240–4249. [Google Scholar] [CrossRef]
- Cui, S.W.; Mazza, G.; Biliaderis, C.G. Chemical structure, molecular size distributions, and rheological properties of flaxseed gum. J. Agric. Food Chem. 1994, 42, 1891–1895. [Google Scholar] [CrossRef]
- Maukonen, J.; Saarela, M. Human gut microbiota: Does diet matter? Proc. Nutr. Soc. 2015, 74, 23–36. [Google Scholar] [CrossRef]
- Broekaert, W.F.; Courtin, C.M.; Verbeke, K.; Van de Wiele, T.; Verstraete, W.; Delcour, J.A. Prebiotic and other health-related effects of cereal-derived arabinoxylans, arabinoxylan-oligosaccharides, and xylooligosaccharides. Crit. Rev. Food Sci. Nutr. 2011, 512, 178–194. [Google Scholar] [CrossRef] [PubMed]
- Verkhnyatskaya, S.; Ferrari, M.; de Vos, P.; Walvoort, M.T. Shaping the infant microbiome with non-digestible carbohydrates. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Rivière, A.; Moens, F.; Selak, M.; Maes, D.; Weckx, S.; De Vuyst, L. The ability of bifidobacteria to degrade arabinoxylan oligosaccharide constituents and derived oligosaccharides is strain dependent. Appl. Environ. Microbiol. 2014, 80, 204–217. [Google Scholar] [CrossRef] [PubMed]
- Rivière, A.; Selak, M.; Geirnaert, A.; Van den Abbeele, P.; De Vuyst, L. Complementary mechanisms for degradation of inulin-type fructans and arabinoxylan oligosaccharides among bifidobacterial strains suggest bacterial cooperation. Appl. Environ. Microbiol. 2018, 84, e02893-17. [Google Scholar] [CrossRef]
- De Paepe, K.; Kerckhof, F.M.; Verspreet, J.; Courtin, C.M.; Van de Wiele, T. Inter-individual differences determine the outcome of wheat bran colonization by the human gut microbiome. Environ. Microbiol. 2017, 19, 3251–3267. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, K.; Verspreet, J.; Verbeke, K.; Raes, J.; Courtin, C.M.; Van de Wiele, T. Introducing insoluble wheat bran as a gut microbiota niche in an in vitro dynamic gut model stimulates propionate and butyrate production and induces colon region specific shifts in the luminal and mucosal microbial community. Environ. Microbiol. 2018, 20, 3406–3426. [Google Scholar] [CrossRef]
- Moczkowska, M.; Karp, S.; Niu, Y.; Kurek, M.A. Enzymatic, enzymatic-ultrasonic and alkaline extraction of soluble dietary fibre from flaxseed—A physicochemical approach. Food Hydrocoll. 2019, 90, 105–112. [Google Scholar] [CrossRef]
- Louis, P.; Duncan, S.H.; McCrae, S.I.; Millar, J.; Jackson, M.S.; Flint, H.J. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacterial. 2004, 186, 2099–2106. [Google Scholar] [CrossRef]
- Kuehn, J.; Ismael, Z.; Long, P.F.; Barker, C.I.; Sharland, M. Reported rates of diarrhea following oral penicillin therapy in pediatric clinical trials. J. Pediatr. Pharmacol. Ther. 2015, 20, 90–104. [Google Scholar]
- Mathur, S.; Fuchs, A.; Bielicki, J.; Van Den Anker, J.; Sharland, M. Antibiotic use for community-acquired pneumonia in neonates and children. WHO Evid. Rev. Paediatr. Int. Child Health 2018, 38 (Suppl. 1), S66–S75. [Google Scholar] [CrossRef]
- Mangin, I.; Suau, A.; Gotteland, M.; Brunser, O.; Pochart, P. Amoxicillin treatment modifies the composition of Bifidobacterium species in infant intestinal microbiota. Anaerobe 2010, 16, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Mangin, I.; Lévêque, C.; Magne, F.; Suau, A.; Pochart, P. Long-term changes in human colonic Bifidobacterium populations induced by a 5-day oral amoxicillin-clavulanic acid treatment. PLoS ONE 2012, 7, e50257. [Google Scholar] [CrossRef]
- MacPherson, C.W.; Mathieu, O.; Tremblay, J.; Champagne, J.; Nantel, A.; Girard, S.A.; Tompkins, T.A. Gut bacterial microbiota and its resistome rapidly recover to basal state levels after short-term amoxicillin-clavulanic acid treatment in healthy adults. Sci. Rep. 2018, 8, 11192. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Wu, H.; Wu, S.D.; Lu, N.; Wang, Y.T.; Liu, H.N.; Shen, X.Z. Parasutterella, in association with irritable bowel syndrome and intestinal chronic inflammation. J. Gastroenterol. Hepatol. 2018, 33, 1844–1852. [Google Scholar] [CrossRef]
- Sibakov, J.; Myllymäki, O.; Suortti, T.; Kaukovirta-Norja, A.; Lehtinen, P.; Poutanen, K. Comparison of acid and enzymatic hydrolyses of oat bran β-glucan at low water content. Food Res. Int. 2013, 52, 99–108. [Google Scholar] [CrossRef]
- Maathuis, A.; Hoffman, A.; Evans, A.; Sanders, L.; Venema, K. The effect of the undigested fraction of maize products on the activity and composition of the microbiota determined in a dynamic in vitro model of the human proximal large intestine. J. Am. Coll. Nutr. 2009, 28, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Bernalier-Donadille, A. The cellulolytic microflora of the human colon: Evidence of microcrystalline cellulose-degrading bacteria in methane-excreting subjects. FEMS Microbial. Ecol. 2003, 46, 81–89. [Google Scholar] [CrossRef]
- Tannergren, C.; Bergendal, A.; Lennernäs, H.; Abrahamsson, B. Toward an increased understanding of the barriers to colonic drug absorption in humans: Implications for early controlled release candidate assessment. Mol. Pharm. 2009, 6, 60–73. [Google Scholar] [CrossRef]
- Penn, R.; Ward, B.J.; Strande, L.; Maurer, M. Review of synthetic human faeces and faecal sludge for sanitation and wastewater research. Water Res. 2018, 132, 222–240. [Google Scholar] [CrossRef]
- Simões, C.D.; Maukonen, J.; Kaprio, J.; Rissanen, A.; Pietiläinen, K.H.; Saarela, M. Habitual dietary intake is associated with stool microbiota composition in monozygotic twins. J. Nutr. 2013, 143, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Development of a semiquantitative degenerate real-time PCR-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl. Environ. Microbiol. 2007, 73, 2009–2012. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-spurce, platform-independent, community supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]




| Sample 1 | Dietary Fiber 1, % (as is) | Protein 2, % | Digestible Starch and Available Carbohydrates (% as Is) 2 | |||
|---|---|---|---|---|---|---|
| Insoluble 2 | Soluble 2 | Oligos 3 | Total 4 | |||
| LS | 25.7 ± 0.1 | 14.9 ± 0.1 | 1.4 | 42.0 | 29.7 ± 0.1 | 0.1 ± 0.1 |
| RB | 35.5 ± 0.1 | 6.7 ± 0.0 | 7.0 | 49.2 | 15.2 ± 0.2 | 14.6 ± 0.3 |
| EnzRB | 25.3 ± 0.3 | 4.9 ± 0.2 | 11.4 | 41.6 | 14.8 ± 0.3 | 16.8 ± 0.1 |
| Sol EnzRB | 0.0 ± 0.0 | 6.6 ± 0.8 | 25.5 | 32.1 | 11.1 ± 0.1 | 28.1 ± 0.2 |
| Insol EnzRB | 37.1 ± 0.3 | 6.2 ± 0.0 | 8.7 | 51.9 | 16.3 ± 0.0 | 12.9 ± 0.2 |
| SolBG | 1.1 ± 1.1 | 52.3 ± 0.8 | 6.4 | 59.8 | 11.6 ± 0.3 | 11.9 ± 0.0 |
| RF:BG:LS | 16.9 ± 0.6 | 25.3 ± 0.3 | 4.9 | 47.1 | 15.5 ± 0.1 | 12.0 ± 0.0 |
| Fiber | Short Name | Supplier | Product | Specifications |
|---|---|---|---|---|
| Microcrystalline Cellulose | MCC | JRS Pharma | VIVAPUR® 105 | Average particle size by laser diffraction 15 µm |
| Resistant Starch | RS | Ingredion | HI-MAIZE® 260 | Dietary fiber 56%, other carbohydrates 31% |
| Xylo-oligosaccharide | XOS | Shandong Longlive Bio-Technology | Xylo-oligosaccharide | Purity >95% |
| Inulin | Inulin | Beneo | Orafti® HSI | Inulin content ~88% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsitko, I.; Wiik-Miettinen, F.; Mattila, O.; Rosa-Sibakov, N.; Seppänen-Laakso, T.; Maukonen, J.; Nordlund, E.; Saarela, M. A Small In Vitro Fermentation Model for Screening the Gut Microbiota Effects of Different Fiber Preparations. Int. J. Mol. Sci. 2019, 20, 1925. https://doi.org/10.3390/ijms20081925
Tsitko I, Wiik-Miettinen F, Mattila O, Rosa-Sibakov N, Seppänen-Laakso T, Maukonen J, Nordlund E, Saarela M. A Small In Vitro Fermentation Model for Screening the Gut Microbiota Effects of Different Fiber Preparations. International Journal of Molecular Sciences. 2019; 20(8):1925. https://doi.org/10.3390/ijms20081925
Chicago/Turabian StyleTsitko, Irina, Fanny Wiik-Miettinen, Outi Mattila, Natalia Rosa-Sibakov, Tuulikki Seppänen-Laakso, Johanna Maukonen, Emilia Nordlund, and Maria Saarela. 2019. "A Small In Vitro Fermentation Model for Screening the Gut Microbiota Effects of Different Fiber Preparations" International Journal of Molecular Sciences 20, no. 8: 1925. https://doi.org/10.3390/ijms20081925
APA StyleTsitko, I., Wiik-Miettinen, F., Mattila, O., Rosa-Sibakov, N., Seppänen-Laakso, T., Maukonen, J., Nordlund, E., & Saarela, M. (2019). A Small In Vitro Fermentation Model for Screening the Gut Microbiota Effects of Different Fiber Preparations. International Journal of Molecular Sciences, 20(8), 1925. https://doi.org/10.3390/ijms20081925