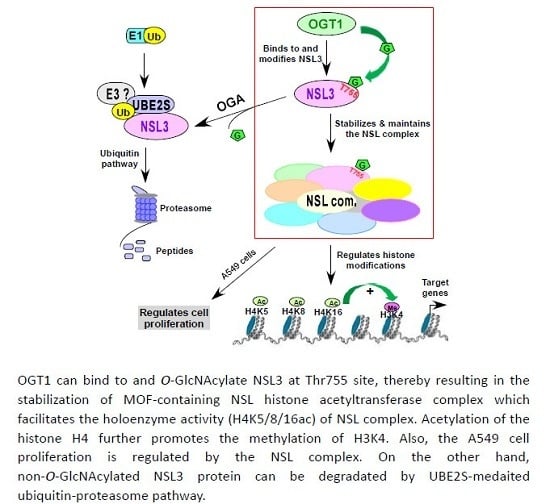
O-GlcNAc-Modification of NSL3 at Thr755 Site Maintains the Holoenzyme Activity of MOF/NSL Histone Acetyltransferase Complex
Abstract
:1. Introduction
2. Results
2.1. OGT1 Stabilized NSL3 through O-GlcNAcylating Its C-Terminus
2.2. Ubiquitin-Conjugating Enzyme UBE2S, But Not UBE2N, Directly Bound to NSL3
2.3. O-GlcNAcylation of NSL3 by OGT1 Was Tightly Associated with UBE2S-Mediated Ubiquitin-Degradation Pathway
2.4. O-GlcNAc-Modification of NSL3 by OGT1 Mainly Occurred at Thr755
2.5. O-GlcNAcylation of NSL3 Thr755 Site by OGT1 Was Connected to the NSL3 Stability
2.6. O-GlcNAcylation of NSL3 at Thr755 Site Was Required for Maintaining the Integrity of NSL Complex and Its Holoenzyme Activity
3. Discussion
4. Materials and Methods
4.1. Antibodies
4.2. Cell Culture
4.3. Plasmids
4.4. Expression of Recombinant Proteins in Sf21 Insect Cells
4.5. In Vitro O-GlcNAc Transferase Assay
4.6. Transient Transfection and Immunoprecipitation (IP)
4.7. Immunofluorescence Staining
4.8. Colony Formation Experiment
4.9. Mass Spectrometry
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| OGT | O-linked N-acetylglucosamine transferase |
| O-GlcNAc | O-linked N-acetylglucosamine |
| NSL | Nonspecific lethal |
| OGA | O-GlcNAcase |
| HAT MOF | Histone acetyltransferase Males absent on the first |
References
- Wells, L.; Hart, G.W. O-GlcNAc turns twenty: Functional implications for post-translational modification of nuclear and cytosolic proteins with a sugar. FEBS Lett. 2003, 546, 154–158. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wells, L.; Comer, F.I.; Parker, G.J.; Hart, G.W. Dynamic O-glycosylation of nuclear and cytosolic proteins: Cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J. Biol. Chem. 2001, 276, 9838–9845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mailleux, F.; Gélinas, R.; Beauloye, C.; Horman, S.; Bertrand, L. O-GlcNAcylation, enemy or ally during cardiac hypertrophy development? Biochim. Biophys. Acta 2016, 1862, 2232–2243. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.A. O-GlcNAcylation at Promoters, Nutrient Sensors, and Transcriptional Regulation. Biochim. Biophys. Acta 2013, 1829, 12021–12206. [Google Scholar] [CrossRef] [Green Version]
- Jiang, M.; Qiu, Z.; Zhang, S.; Fan, X.; Cai, X.; Xu, B.; Li, X.; Zhou, J.; Zhang, X.; Chu, Y.; et al. Elevated O-GlcNAcylation promotes gastric cancer cells proliferation by modulating cell cycle related proteins and ERK 1/2 signaling. Oncotarget 2016, 7, 61390–61402. [Google Scholar] [CrossRef] [Green Version]
- Geng, F.; Zhu, W.; Anderson, R.A.; Leber, B.; Andrews, D.W. Multiple post-translational modifications regulate E-cadherin transport during apoptosis. J. Cell Sci. 2012, 125, 2615–2625. [Google Scholar] [CrossRef] [Green Version]
- Hardivillé, S.; Hart, G.W. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014, 20, 208–213. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.H.; Kim, J.E.; Nam, H.W.; Ju, J.W.; Kim, H.S.; Kim, Y.S.; Cho, J.W. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 2006, 8, 1074–1083. [Google Scholar] [CrossRef]
- Wells, L.; Vosseller, K.; Hart, G.W. A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell Mol. Life Sci. 2003, 60, 222–228. [Google Scholar] [CrossRef]
- Slawson, C.; Lakshmanan, T.; Knapp, S.; Hart, G.W. A Mitotic GlcNAcylation/Phosphorylation Signaling Complex Alters the Posttranslational State of the Cytoskeletal Protein Vimentin. Mol. Biol. Cell 2008, 19, 4130–4140. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Zhao, L.; Feng, Z.; Yu, C.; Ding, J.; Wang, L.; Wang, F.; Liu, D.; Zhu, H.; Xing, F.; et al. O-Linked N-acetylglucosamine transferase 1 regulates global histone H4 acetylation via stabilization of the nonspecific lethal protein NSL. J. Biol. Chem. 2017, 292, 10014–10025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Shah, J.A.; Cai, Y.; Jin, J. ‘O-GlcNAc Code’ Mediated Biological Functions of Downstream Proteins. Molecules 2018, 23, 1967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, C.; Gu, Y.; Shan, H.; Mi, W.; Sun, J.; Shi, M.; Zhang, X.; Lu, X.; Han, F.; Gong, Q.; et al. O-GlcNAcylation of SIRT1 enhances its deacetylase activity and promotes cytoprotection under stress. Nat. Commun. 2017, 8, 1491. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Zhu, X.; He, W.; Zhao, S.; Zhao, W.; Zhu, C. Resveratrol inhibits the hydrogen dioxide-induced apoptosis via Sirt 1 activation in osteoblast cells. Biosci. Biotechnol. Biochem. 2015, 79, 1779–1786. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Zhang, L.; Zhou, C.; Lin, N.; Liu, A. Sirt 1 activator inhibits the AGE-induced apoptosis and p53 acetylation in human vascular endothelial cells. J. Toxicol. Sci. 2015, 40, 615–624. [Google Scholar] [CrossRef] [Green Version]
- Capotosti, F.; Guernier, S.; Lammers, F.; Waridel, P.; Cai, Y.; Jin, J.; Conaway, J.W.; Conaway, R.C.; Herr, W. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 2011, 144, 376–388. [Google Scholar] [CrossRef] [Green Version]
- Ruan, H.B.; Han, X.; Li, M.D.; Singh, J.P.; Qian, K.; Azarhoush, S.; Zhao, L.; Bennett, A.M.; Samuel, V.T.; Wu, J.; et al. O-GlcNAc Transferase/Host Cell Factor C1 Complex Regulates Gluconeogenesis by Modulating PGC-1α Stability. Cell Metab. 2012, 16, 226–237. [Google Scholar] [CrossRef] [Green Version]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [Green Version]
- Deplus, R.; Delatte, B.; Schwinn, M.K.; Defrance, M.; Méndez, J.; Murphy, N.; Dawson, M.A.; Volkmar, M.; Putmans, P.; Calonne, E.; et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013, 32, 645–655. [Google Scholar] [CrossRef]
- Cai, Y.; Jin, J.; Swanson, S.K.; Cole, M.D.; Choi, S.H.; Florens, L.; Washburn, M.P.; Conaway, J.W.; Conaway, R.C. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal(MSL)complex. J. Biol. Chem. 2010, 285, 4268–4272. [Google Scholar] [CrossRef] [Green Version]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turek, I.; Tischer, N.; Lassig, R.; Trujillo, M. Multi-tiered pairing selectivity between E2 ubiquitin-conjugating enzymes and E3 ligases. J. Biol. Chem. 2018, 42, 63241–66336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liess, A.K.L.; Kucerova, A.; Schweimer, K.; Yu, L.; Roumeliotis, T.; Diebold, M.; Dybkov, O.; Sotriffer, C.; Urlaub, H.; Choudhary, J.S.; et al. Autoinhibition Mechanism of the Ubiquitin-Conjugating Enzyme UBE2S by Autoubiquitination. Structure 2019, 27, 1195–1210.e7. [Google Scholar] [CrossRef] [PubMed]
- Scaglione, K.M.; Basrur, V.; Ashraf, N.S.; Konen, J.R.; Elenitoba-Johnson, K.S.; Todi, S.V.; Paulson, H.L. The ubiquitin-conjugating enzyme (E2) Ube2w ubiquitinates the N terminus of substrates. J. Biol. Chem. 2013, 288, 187841–188788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiki, R.; Hashiba, W.; Sekine, H.; Yokoyama, A.; Chikanishi, T.; Ito, S.; Imai, Y.; Kim, J.; He, H.H.; Igarashi, K.; et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 2011, 480, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Jiang, W.; Zhou, P.; Liu, L.; Wan, X.; Yuan, X.; Wang, X.; Chen, M.; Chen, J.; Yang, J.; et al. Mixed Lineage Leukemia 5 (MLL5) Protein Stability Is Cooperatively Regulated by O-GlcNac Transferase (OGT) and Ubiquitin Specific Protease 7 (USP7). PLoS ONE 2015, 10, e0145023. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.S.; Lo, P.W.; Yeh, Y.H.; Hsu, P.H.; Peng, S.H.; Teng, Y.C.; Kang, M.L.; Wong, C.H.; Juan, L.J. O-GlcNAcylation regulates EZH2 protein stability and function. Proc. Natl. Acad. Sci. USA 2014, 111, 1355–1360. [Google Scholar] [CrossRef] [Green Version]
- Ferrer, C.M.; Lu, T.Y.; Bacigalupa, Z.A.; Katsetos, C.D.; Sinclair, D.A.; Reginato, M.J. O-GlcNAcylation regulates breast cancer metastasis via SIRT1 modulation of FOXM1 pathway. Oncogene 2017, 36, 559–569. [Google Scholar] [CrossRef] [Green Version]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Li, M.; Wei, T.; Feng, C.; Wu, T.; Shah, J.A.; Liu, H.; Wang, F.; Cai, Y.; Jin, J. O-GlcNAc-Modification of NSL3 at Thr755 Site Maintains the Holoenzyme Activity of MOF/NSL Histone Acetyltransferase Complex. Int. J. Mol. Sci. 2020, 21, 173. https://doi.org/10.3390/ijms21010173
Zhao L, Li M, Wei T, Feng C, Wu T, Shah JA, Liu H, Wang F, Cai Y, Jin J. O-GlcNAc-Modification of NSL3 at Thr755 Site Maintains the Holoenzyme Activity of MOF/NSL Histone Acetyltransferase Complex. International Journal of Molecular Sciences. 2020; 21(1):173. https://doi.org/10.3390/ijms21010173
Chicago/Turabian StyleZhao, Linhong, Min Li, Tao Wei, Chang Feng, Tingting Wu, Junaid Ali Shah, Hongsen Liu, Fei Wang, Yong Cai, and Jingji Jin. 2020. "O-GlcNAc-Modification of NSL3 at Thr755 Site Maintains the Holoenzyme Activity of MOF/NSL Histone Acetyltransferase Complex" International Journal of Molecular Sciences 21, no. 1: 173. https://doi.org/10.3390/ijms21010173
APA StyleZhao, L., Li, M., Wei, T., Feng, C., Wu, T., Shah, J. A., Liu, H., Wang, F., Cai, Y., & Jin, J. (2020). O-GlcNAc-Modification of NSL3 at Thr755 Site Maintains the Holoenzyme Activity of MOF/NSL Histone Acetyltransferase Complex. International Journal of Molecular Sciences, 21(1), 173. https://doi.org/10.3390/ijms21010173