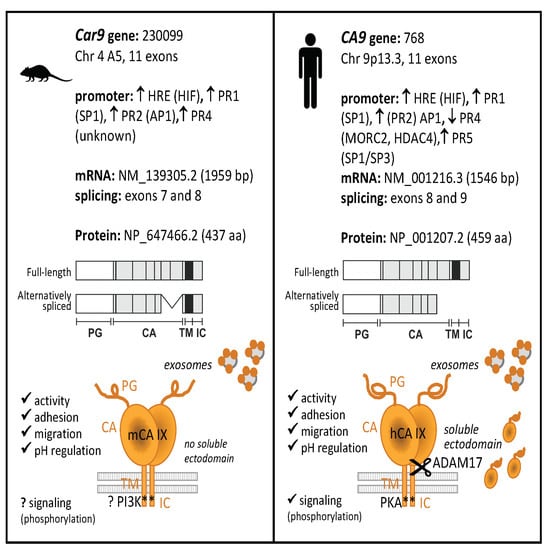
Carbonic Anhydrase IX—Mouse versus Human
Abstract
:1. Introduction
2. Results
2.1. Transcriptional and Post-Transcriptional Regulation of the Car9 Gene
2.1.1. In Silico Analysis of the Mouse Car9 Promoter Sequence
2.1.2. Hypoxia- and Density-Induced Activity of the Mouse Car9 Promoter
2.1.3. The role of SP1 and AP1-Binding Sites in the Transcriptional Activity of Car9
2.1.4. Significant Differences Observed within PR4 and PR5 Regions
2.2. Characterization of the Mouse CA IX Protein
2.2.1. Functional Analyses of mCA IX
2.2.2. Extracellular Forms of mCA IX—mCA IX Ectodomain Shedding
2.2.3. Extracellular Forms of mCA IX—Presence of CA IX in Exosomes Derived from Murine Cells
2.3. Expression of mCA IX
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Plasmids
4.3. Transient Transfection and Luciferase Assay
4.4. Immunofluorescence
4.5. Cell Dissociation Assay
4.6. Real-Time Monitoring of Migration with the xCELLigence System
4.7. Isolation of Exosomes
4.8. Nanoparticle Tracking Analysis
4.9. ELISA
4.10. Western Blotting Analysis
4.11. Shedding
4.12. Bioinformatics
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Statement
Abbreviations
| AS | Alternative splicing |
| CA | Carbonic anhydrase |
| CA IX | Carbonic anhydrase IX |
| hCA IX | Human carbonic anhydrase IX |
| mCA IX | Mouse carbonic anhydrase IX |
| ECD | Extracellular domain |
| ELISA | Enzyme-linked Immunosorbent Assay |
| FL | Full length |
| HIF | Hypoxia-inducible factor |
| HRE | Hypoxia-responsive element |
| IC | Intracytoplasmic |
| IHC | Immunohistochemistry |
| mCA IX | Mouse carbonic anhydrase IX |
| PG | Proteoglycan |
| PKA | Protein kinase A |
| PMA | Phorbol-12-myristate-13-acetate |
| PR | Protected region |
| TF | Transcription factor |
| TM | Transmembrane |
| TSS | Transcription start site |
References
- Hammond, E.M.; Asselin, M.C.; Forster, D.; O’Connor, J.P.; Senra, J.M.; Williams, K.J. The meaning, measurement and modification of hypoxia in the laboratory and the clinic. Clin. Oncol. (R. Coll. Radiol.) 2014, 26, 277–288. [Google Scholar] [CrossRef]
- Semenza, G.L. Hydroxylation of HIF-1: Oxygen sensing at the molecular level. Physiology (Bethesda) 2004, 19, 176–182. [Google Scholar] [CrossRef] [Green Version]
- Pastorekova, S.; Parkkila, S.; Parkkila, A.K.; Opavsky, R.; Zelnik, V.; Saarnio, J.; Pastorek, J. Carbonic anhydrase IX, MN/CA IX: Analysis of stomach complementary DNA sequence and expression in human and rat alimentary tracts. Gastroenterology 1997, 112, 398–408. [Google Scholar] [CrossRef]
- Svastova, E.; Hulikova, A.; Rafajova, M.; Zat’ovicova, M.; Gibadulinova, A.; Casini, A.; Cecchi, A.; Scozzafava, A.; Supuran, C.T.; Pastorek, J.; et al. Hypoxia activates the capacity of tumor-associated carbonic anhydrase IX to acidify extracellular pH. FEBS Lett. 2004, 577, 439–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swietach, P.; Patiar, S.; Supuran, C.T.; Harris, A.L.; Vaughan-Jones, R.D. The role of carbonic anhydrase 9 in regulating extracellular and intracellular ph in three-dimensional tumor cell growths. J. Biol. Chem. 2009, 284, 20299–20310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svastova, E.; Witarski, W.; Csaderova, L.; Kosik, I.; Skvarkova, L.; Hulikova, A.; Zatovicova, M.; Barathova, M.; Kopacek, J.; Pastorek, J.; et al. Carbonic anhydrase IX interacts with bicarbonate transporters in lamellipodia and increases cell migration via its catalytic domain. J. Biol. Chem. 2012, 287, 3392–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swayampakula, M.; McDonald, P.C.; Vallejo, M.; Coyaud, E.; Chafe, S.C.; Westerback, A.; Venkateswaran, G.; Shankar, J.; Gao, G.; Laurent, E.M.N.; et al. The interactome of metabolic enzyme carbonic anhydrase IX reveals novel roles in tumor cell migration and invadopodia/MMP14-mediated invasion. Oncogene 2017, 36, 6244–6261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; McIntyre, D.; Honess, D.; Hulikova, A.; Pacheco-Torres, J.; Cerdan, S.; Swietach, P.; Harris, A.L.; Griffiths, J.R. Carbonic anhydrase IX is a pH-stat that sets an acidic tumour extracellular pH in vivo. Br. J. Cancer 2018, 119, 622–630. [Google Scholar] [CrossRef] [Green Version]
- Beasley, N.J.; Wykoff, C.C.; Watson, P.H.; Leek, R.; Turley, H.; Gatter, K.; Pastorek, J.; Cox, G.J.; Ratcliffe, P.; Harris, A.L. Carbonic anhydrase IX, an endogenous hypoxia marker, expression in head and neck squamous cell carcinoma and its relationship to hypoxia, necrosis, and microvessel density. Cancer Res. 2001, 61, 5262–5267. [Google Scholar]
- Chia, S.K.; Wykoff, C.C.; Watson, P.H.; Han, C.; Leek, R.D.; Pastorek, J.; Gatter, K.C.; Ratcliffe, P.; Harris, A.L. Prognostic significance of a novel hypoxia-regulated marker, carbonic anhydrase IX, in invasive breast carcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2001, 19, 3660–3668. [Google Scholar] [CrossRef]
- Koukourakis, M.I.; Giatromanolaki, A.; Sivridis, E.; Simopoulos, K.; Pastorek, J.; Wykoff, C.C.; Gatter, K.C.; Harris, A.L. Hypoxia-regulated carbonic anhydrase-9 (CA9) relates to poor vascularization and resistance of squamous cell head and neck cancer to chemoradiotherapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2001, 7, 3399–3403. [Google Scholar]
- Haapasalo, J.A.; Nordfors, K.M.; Hilvo, M.; Rantala, I.J.; Soini, Y.; Parkkila, A.K.; Pastorekova, S.; Pastorek, J.; Parkkila, S.M.; Haapasalo, H.K. Expression of carbonic anhydrase IX in astrocytic tumors predicts poor prognosis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 473–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Driessen, A.; Landuyt, W.; Pastorekova, S.; Moons, J.; Goethals, L.; Haustermans, K.; Nafteux, P.; Penninckx, F.; Geboes, K.; Lerut, T.; et al. Expression of carbonic anhydrase IX (CA IX), a hypoxia-related protein, rather than vascular-endothelial growth factor (VEGF), a pro-angiogenic factor, correlates with an extremely poor prognosis in esophageal and gastric adenocarcinomas. Ann. Surg. 2006, 243, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Pastorek, J.; Pastorekova, S. Hypoxia-induced carbonic anhydrase IX as a target for cancer therapy: From biology to clinical use. Semin. Cancer Biol. 2015, 31, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Hilvo, M.; Rafajova, M.; Pastorekova, S.; Pastorek, J.; Parkkila, S. Expression of carbonic anhydrase IX in mouse tissues. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2004, 52, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Gut, M.O.; Parkkila, S.; Vernerova, Z.; Rohde, E.; Zavada, J.; Hocker, M.; Pastorek, J.; Karttunen, T.; Gibadulinova, A.; Zavadova, Z.; et al. Gastric hyperplasia in mice with targeted disruption of the carbonic anhydrase gene Car9. Gastroenterology 2002, 123, 1889–1903. [Google Scholar] [CrossRef] [PubMed]
- Kaluz, S.; Kaluzova, M.; Opavsky, R.; Pastorekova, S.; Gibadulinova, A.; Dequiedt, F.; Kettmann, R.; Pastorek, J. Transcriptional regulation of the MN/CA 9 gene coding for the tumor-associated carbonic anhydrase IX. Identification and characterization of a proximal silencer element. J. Biol. Chem. 1999, 274, 32588–32595. [Google Scholar] [CrossRef] [Green Version]
- Takacova, M.; Barathova, M.; Hulikova, A.; Ohradanova, A.; Kopacek, J.; Parkkila, S.; Pastorek, J.; Pastorekova, S.; Zatovicova, M. Hypoxia-inducible expression of the mouse carbonic anhydrase IX demonstrated by new monoclonal antibodies. Int. J. Oncol. 2007, 31, 1103–1110. [Google Scholar]
- Wykoff, C.C.; Beasley, N.J.; Watson, P.H.; Turner, K.J.; Pastorek, J.; Sibtain, A.; Wilson, G.D.; Turley, H.; Talks, K.L.; Maxwell, P.H.; et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000, 60, 7075–7083. [Google Scholar]
- Kaluz, S.; Kaluzova, M.; Stanbridge, E.J. Expression of the hypoxia marker carbonic anhydrase IX is critically dependent on SP1 activity. Identification of a novel type of hypoxia-responsive enhancer. Cancer Res. 2003, 63, 917–922. [Google Scholar]
- Kaluzova, M.; Pastorekova, S.; Svastova, E.; Pastorek, J.; Stanbridge, E.J.; Kaluz, S. Characterization of the MN/CA 9 promoter proximal region: A role for specificity protein (SP) and activator protein 1 (AP1) factors. Biochem. J. 2001, 359, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Svastova, E.; Zilka, N.; Zat’ovicova, M.; Gibadulinova, A.; Ciampor, F.; Pastorek, J.; Pastorekova, S. Carbonic anhydrase IX reduces E-cadherin-mediated adhesion of MDCK cells via interaction with beta-catenin. Exp. Cell Res. 2003, 290, 332–345. [Google Scholar] [CrossRef]
- Zavada, J.; Zavadova, Z.; Zat’ovicova, M.; Hyrsl, L.; Kawaciuk, I. Soluble form of carbonic anhydrase IX (CA IX) in the serum and urine of renal carcinoma patients. Br. J. Cancer 2003, 89, 1067–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zatovicova, M.; Sedlakova, O.; Svastova, E.; Ohradanova, A.; Ciampor, F.; Arribas, J.; Pastorek, J.; Pastorekova, S. Ectodomain shedding of the hypoxia-induced carbonic anhydrase IX is a metalloprotease-dependent process regulated by TACE/ADAM17. Br. J. Cancer 2005, 93, 1267–1276. [Google Scholar] [CrossRef] [Green Version]
- Horie, K.; Kawakami, K.; Fujita, Y.; Sugaya, M.; Kameyama, K.; Mizutani, K.; Deguchi, T.; Ito, M. Exosomes expressing carbonic anhydrase 9 promote angiogenesis. Biochem. Biophys. Res. Commun. 2017, 492, 356–361. [Google Scholar] [CrossRef]
- Logozzi, M.; Capasso, C.; Di Raimo, R.; Del Prete, S.; Mizzoni, D.; Falchi, M.; Supuran, C.T.; Fais, S. Prostate cancer cells and exosomes in acidic condition show increased carbonic anhydrase IX expression and activity. J. Enzym. Inhib. Med. Chem. 2019, 34, 272–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zatovicova, M.; Pastorekova, S. Modulation of cell surface density of carbonic anhydrase IX by shedding of the ectodomain and endocytosis. Acta Virol. 2013, 57, 257–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, Y.; Li, Y.; Zhang, J.; Liu, D.; Liu, F.; Zhao, Y.; Shen, T.; Li, F. Involvement of histone deacetylation in MORC2-mediated down-regulation of carbonic anhydrase IX. Nucleic Acids Res. 2010, 38, 2813–2824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cadigan, K.M.; Waterman, M.L. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef]
- Barathova, M.; Takacova, M.; Holotnakova, T.; Gibadulinova, A.; Ohradanova, A.; Zatovicova, M.; Hulikova, A.; Kopacek, J.; Parkkila, S.; Supuran, C.T.; et al. Alternative splicing variant of the hypoxia marker carbonic anhydrase IX expressed independently of hypoxia and tumour phenotype. Br. J. Cancer 2008, 98, 129–136. [Google Scholar] [CrossRef] [Green Version]
- Hynninen, P.; Hamalainen, J.M.; Pastorekova, S.; Pastorek, J.; Waheed, A.; Sly, W.S.; Tomas, E.; Kirkinen, P.; Parkkila, S. Transmembrane carbonic anhydrase isozymes IX and XII in the female mouse reproductive organs. Reprod. Biol. Endocrinol. 2004, 2, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leppilampi, M.; Karttunen, T.J.; Kivela, J.; Gut, M.O.; Pastorekova, S.; Pastorek, J.; Parkkila, S. Gastric pit cell hyperplasia and glandular atrophy in carbonic anhydrase IX knockout mice: Studies on two strains C57/BL6 and BALB/C. Transgenic Res. 2005, 14, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Kallio, H.; Pastorekova, S.; Pastorek, J.; Waheed, A.; Sly, W.S.; Mannisto, S.; Heikinheimo, M.; Parkkila, S. Expression of carbonic anhydrases IX and XII during mouse embryonic development. BMC Dev. Biol. 2006, 6, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, P.; Leppilampi, M.; Pastorekova, S.; Pastorek, J.; Waheed, A.; Sly, W.S.; Parkkila, S. Carbonic anhydrase gene expression in CA II-deficient (Car2-/-) and CA IX-deficient (Car9-/-) mice. J. Physiol. 2006, 571, 319–327. [Google Scholar] [CrossRef]
- Scheibe, R.J.; Gros, G.; Parkkila, S.; Waheed, A.; Grubb, J.H.; Shah, G.N.; Sly, W.S.; Wetzel, P. Expression of membrane-bound carbonic anhydrases IV, IX, and XIV in the mouse heart. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2006, 54, 1379–1391. [Google Scholar] [CrossRef] [Green Version]
- Scheibe, R.J.; Mundhenk, K.; Becker, T.; Hallerdei, J.; Waheed, A.; Shah, G.N.; Sly, W.S.; Gros, G.; Wetzel, P. Carbonic anhydrases IV and IX: Subcellular localization and functional role in mouse skeletal muscle. Am. J. Physiol. Cell Physiol. 2008, 294, C402–C412. [Google Scholar] [CrossRef]
- Li, T.; Liu, X.; Riederer, B.; Nikolovska, K.; Singh, A.K.; Makela, K.A.; Seidler, A.; Liu, Y.; Gros, G.; Bartels, H.; et al. Genetic ablation of carbonic anhydrase IX disrupts gastric barrier function via claudin-18 downregulation and acid backflux. Acta Physiol. 2018, 222, e12923. [Google Scholar] [CrossRef] [Green Version]
- Kallio, H.; Hilvo, M.; Rodriguez, A.; Lappalainen, E.H.; Lappalainen, A.M.; Parkkila, S. Global transcriptional response to carbonic anhydrase IX deficiency in the mouse stomach. BMC Genom. 2010, 11, 397. [Google Scholar] [CrossRef] [Green Version]
- Parks, S.K.; Chiche, J.; Pouyssegur, J. Disrupting proton dynamics and energy metabolism for cancer therapy. Nat. Rev. Cancer 2013, 13, 611–623. [Google Scholar] [CrossRef]
- Radvak, P.; Repic, M.; Svastova, E.; Takacova, M.; Csaderova, L.; Strnad, H.; Pastorek, J.; Pastorekova, S.; Kopacek, J. Suppression of carbonic anhydrase IX leads to aberrant focal adhesion and decreased invasion of tumor cells. Oncol. Rep. 2013, 29, 1147–1153. [Google Scholar] [CrossRef] [Green Version]
- Ward, C.; Meehan, J.; Mullen, P.; Supuran, C.; Dixon, J.M.; Thomas, J.S.; Winum, J.Y.; Lambin, P.; Dubois, L.; Pavathaneni, N.K.; et al. Evaluation of carbonic anhydrase IX as a therapeutic target for inhibition of breast cancer invasion and metastasis using a series of in vitro breast cancer models. Oncotarget 2015, 6, 24856–24870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ditte, P.; Dequiedt, F.; Svastova, E.; Hulikova, A.; Ohradanova-Repic, A.; Zatovicova, M.; Csaderova, L.; Kopacek, J.; Supuran, C.T.; Pastorekova, S.; et al. Phosphorylation of carbonic anhydrase IX controls its ability to mediate extracellular acidification in hypoxic tumors. Cancer Res. 2011, 71, 7558–7567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simko, V.; Iuliano, F.; Sevcikova, A.; Labudova, M.; Barathova, M.; Radvak, P.; Pastorekova, S.; Pastorek, J.; Csaderova, L. Hypoxia induces cancer-associated cAMP/PKA signalling through HIF-mediated transcriptional control of adenylyl cyclases VI and VII. Sci. Rep. 2017, 7, 10121. [Google Scholar] [CrossRef] [PubMed]
- Dorai, T.; Sawczuk, I.S.; Pastorek, J.; Wiernik, P.H.; Dutcher, J.P. The role of carbonic anhydrase IX overexpression in kidney cancer. Eur. J. Cancer 2005, 41, 2935–2947. [Google Scholar] [CrossRef] [PubMed]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Zhou, X.; Yao, Q.; Liu, Y.; Zhang, H.; Dong, Z. HIF-1-mediated production of exosomes during hypoxia is protective in renal tubular cells. Am. J. Physiol. Ren. Physiol. 2017, 313, F906–F913. [Google Scholar] [CrossRef]
- Takacova, M.; Holotnakova, T.; Vondracek, J.; Machala, M.; Pencikova, K.; Gradin, K.; Poellinger, L.; Pastorek, J.; Pastorekova, S.; Kopacek, J. Role of aryl hydrocarbon receptor in modulation of the expression of the hypoxia marker carbonic anhydrase IX. Biochem. J. 2009, 419, 419–425. [Google Scholar] [CrossRef] [Green Version]
- Pastorek, J.; Pastorekova, S.; Callebaut, I.; Mornon, J.P.; Zelnik, V.; Opavsky, R.; Zat’ovicova, M.; Liao, S.; Portetelle, D.; Stanbridge, E.J.; et al. Cloning and characterization of MN, a human tumor-associated protein with a domain homologous to carbonic anhydrase and a putative helix-loop-helix DNA binding segment. Oncogene 1994, 9, 2877–2888. [Google Scholar]
- Pastorekova, S.; Zavadova, Z.; Kostal, M.; Babusikova, O.; Zavada, J. A novel quasi-viral agent, MaTu, is a two-component system. Virology 1992, 187, 620–626. [Google Scholar] [CrossRef]
- Cartharius, K.; Frech, K.; Grote, K.; Klocke, B.; Haltmeier, M.; Klingenhoff, A.; Frisch, M.; Bayerlein, M.; Werner, T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21, 2933–2942. [Google Scholar] [CrossRef] [Green Version]
- Quandt, K.; Frech, K.; Karas, H.; Wingender, E.; Werner, T. MatInd and MatInspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995, 23, 4878–4884. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef] [PubMed]
- Obenauer, J.C.; Cantley, L.C.; Yaffe, M.B. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003, 31, 3635–3641. [Google Scholar] [CrossRef] [Green Version]





| Oligonucleotide | Sequence |
|---|---|
| MPR5Sac | TTTGAGCTCCTATGAACACACCTGCCCCTC |
| MPAXho | TTTCTCGAGAGCTGACTGGGGTGTCCCAGG |
| mHREMs | CCTATTTCCGATGCTTTTACAGCCCGTCCA |
| mHREMa | TGGACGGGCTGTAAAAGCATCGGAAATAGG |
| mSP1Ms | CAAGGCTTGCTCCTAACCTACCCAGCTCCT |
| mSP1Ma | AGGAGCTGGGTAGGTTAGGAGCAAGCCTTG |
| mAP1Ms | AGGGCACTGTGAGTTGGCCTGCTCCCCTCA |
| mAP1Ma | TGAGGGGAGCAGGCCAACTCACAGTGCCCT |
| mPR4s | GGGGAGGGGGCATAGGGCCAGACAGAACCTG |
| mPR4a | CAGGTTCTGTCTGGCCCTATGCCCCCTCCCC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takacova, M.; Barathova, M.; Zatovicova, M.; Golias, T.; Kajanova, I.; Jelenska, L.; Sedlakova, O.; Svastova, E.; Kopacek, J.; Pastorekova, S. Carbonic Anhydrase IX—Mouse versus Human. Int. J. Mol. Sci. 2020, 21, 246. https://doi.org/10.3390/ijms21010246
Takacova M, Barathova M, Zatovicova M, Golias T, Kajanova I, Jelenska L, Sedlakova O, Svastova E, Kopacek J, Pastorekova S. Carbonic Anhydrase IX—Mouse versus Human. International Journal of Molecular Sciences. 2020; 21(1):246. https://doi.org/10.3390/ijms21010246
Chicago/Turabian StyleTakacova, Martina, Monika Barathova, Miriam Zatovicova, Tereza Golias, Ivana Kajanova, Lenka Jelenska, Olga Sedlakova, Eliska Svastova, Juraj Kopacek, and Silvia Pastorekova. 2020. "Carbonic Anhydrase IX—Mouse versus Human" International Journal of Molecular Sciences 21, no. 1: 246. https://doi.org/10.3390/ijms21010246
APA StyleTakacova, M., Barathova, M., Zatovicova, M., Golias, T., Kajanova, I., Jelenska, L., Sedlakova, O., Svastova, E., Kopacek, J., & Pastorekova, S. (2020). Carbonic Anhydrase IX—Mouse versus Human. International Journal of Molecular Sciences, 21(1), 246. https://doi.org/10.3390/ijms21010246