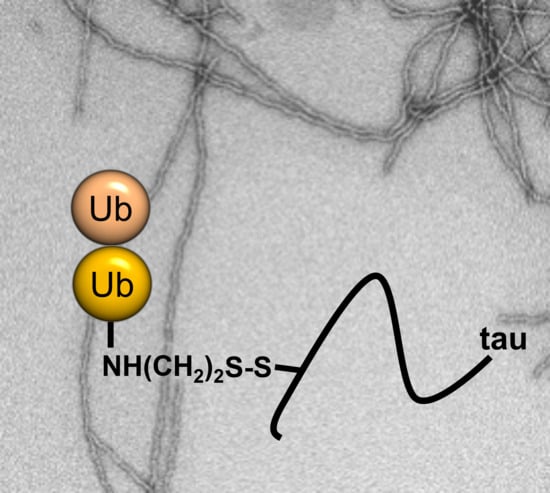
Semisynthetic Modification of Tau Protein with Di-Ubiquitin Chains for Aggregation Studies
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Protein Expression and Purification
4.2.2. Production of Di-Ubiquitin Chains
4.2.3. Disulfide-Coupling Reaction
4.2.4. Thioflavin T Aggregation Assay
4.2.5. TEM Analysis
4.2.6. Dot Blotting
4.2.7. Tubulin Polymerization Assay
4.2.8. Mass Spectrometry
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s Disease |
| UPS | Ubiquitin Proteasome System |
| MBD | Microtubule Binding Domain |
| PHFs | Paired Helical Filaments |
| NFT | Neurofibrillary Tangles |
| DTNB | 5,5′-Dithiobis(2-nitrobenzoic acid) |
| MT | Microtubules |
References
- Varshavsky, A. The Ubiquitin System, Autophagy, and Regulated Protein Degradation. Annu. Rev. Biochem. 2017, 86, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Komander, D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009, 37, 937–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshavsky, A. The Ubiquitin System, an Immense Realm. Annu. Rev. Biochem. 2012, 81, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Dantuma, N.P.; Bott, L.C. The ubiquitin-proteasome system in neurodegenerative diseases: Precipitating factor, yet part of the solution. Front. Mol. Neurosci. 2014, 7. [Google Scholar] [CrossRef] [Green Version]
- Pickart, C.M.; Fushman, D. Polyubiquitin chains: Polymeric protein signals. Curr. Opin. Chem. Biol. 2004, 8, 610–616. [Google Scholar] [CrossRef]
- Chen, R.-H.; Chen, Y.-H.; Huang, T.-Y. Ubiquitin-mediated regulation of autophagy. J. Biomed. Sci. 2019, 26, 80. [Google Scholar] [CrossRef]
- Kulathu, Y.; Komander, D. Atypical ubiquitylation — the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012, 13, 508–523. [Google Scholar] [CrossRef]
- Castañeda, C.A.; Chaturvedi, A.; Camara, C.M.; Curtis, J.E.; Krueger, S.; Fushman, D. Linkage-specific conformational ensembles of non-canonical polyubiquitin chains. Phys. Chem. Chem. Phys. 2016, 18, 5771–5788. [Google Scholar] [CrossRef] [Green Version]
- Varadan, R.; Walker, O.; Pickart, C.; Fushman, D. Structural Properties of Polyubiquitin Chains in Solution. J. Mol. Biol. 2002, 324, 637–647. [Google Scholar] [CrossRef]
- Varadan, R.; Assfalg, M.; Haririnia, A.; Raasi, S.; Pickart, C.; Fushman, D. Solution Conformation of Lys 63 -linked Di-ubiquitin Chain Provides Clues to Functional Diversity of Polyubiquitin Signaling. J. Biol. Chem. 2004, 279, 7055–7063. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Blaser, G.; Horrocks, M.H.; Ruedas-Rama, M.J.; Ibrahim, S.; Zhukov, A.A.; Orte, A.; Klenerman, D.; Jackson, S.E.; Komander, D. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature 2012, 492, 266–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popovic, D.; Vucic, D.; Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014, 20, 1242–1253. [Google Scholar] [CrossRef] [PubMed]
- Sulistio, Y.A.; Heese, K. The Ubiquitin-Proteasome System and Molecular Chaperone Deregulation in Alzheimer’s Disease. Mol. Neurobiol. 2016, 53, 905–931. [Google Scholar] [CrossRef] [PubMed]
- Hegde, A.N.; Smith, S.G.; Duke, L.M.; Pourquoi, A.; Vaz, S. Perturbations of Ubiquitin-Proteasome-Mediated Proteolysis in Aging and Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 324. [Google Scholar] [CrossRef]
- Mori, H.; Kondo, J.; Ihara, Y. Ubiquitin is a component of paired helical filaments in Alzheimer’s disease. Science 1987, 235, 1641–1644. [Google Scholar] [CrossRef]
- Oddo, S. The ubiquitin-proteasome system in Alzheimer’s disease. J. Cell. Mol. Med. 2008, 12, 363–373. [Google Scholar] [CrossRef] [Green Version]
- Keck, S.; Nitsch, R.; Grune, T.; Ullrich, O. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer’s disease: Proteasome inhibition in Alzheimer’s disease. J. Neurochem. 2003, 85, 115–122. [Google Scholar] [CrossRef]
- Wang, Y.; Mandelkow, E. Tau in physiology and pathology. Nat. Rev. Neurosci. 2016, 17, 22–35. [Google Scholar] [CrossRef]
- Mandelkow, E.-M.; Mandelkow, E. Biochemistry and Cell Biology of Tau Protein in Neurofibrillary Degeneration. Cold Spring Harb. Perspect. Med. 2012, 2, a006247. [Google Scholar] [CrossRef]
- Martin, L.; Latypova, X.; Terro, F. Post-translational modifications of tau protein: Implications for Alzheimer’s disease. Neurochem. Int. 2011, 58, 458–471. [Google Scholar] [CrossRef]
- Uversky, V.N. Multitude of binding modes attainable by intrinsically disordered proteins: A portrait gallery of disorder-based complexes. Chem. Soc. Rev. 2011, 40, 1623–1634. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, R.; Munari, F.; Becker, S.; Zweckstetter, M. Interaction of the intermembrane space domain of Tim23 protein with mitochondrial membranes. J. Biol. Chem. 2014, 289, 34620–34626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, M.; Stott, K. Disordered domains in chromatin-binding proteins. Essays Biochem. 2019, 63, 147–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Bergen, M.; Friedhoff, P.; Biernat, J.; Heberle, J.; Mandelkow, E.-M.; Mandelkow, E. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif (306VQIVYK311) forming beta structure. Proc. Natl. Acad. Sci. USA 2000, 97, 5129–5134. [Google Scholar] [CrossRef] [Green Version]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H.W. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef] [Green Version]
- Arakhamia, T.; Lee, C.E.; Carlomagno, Y.; Duong, D.M.; Kundinger, S.R.; Wang, K.; Williams, D.; DeTure, M.; Dickson, D.W.; Cook, C.N.; et al. Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains. Cell 2020, 180, 633–644.e12. [Google Scholar] [CrossRef]
- Morishima-Kawashima, M.; Hasegawa, M.; Takio, K.; Suzuki, M.; Titani, K.; Ihara, Y. Ubiquitin is conjugated with amino-terminally processed tau in paired helical filaments. Neuron 1993, 10, 1151–1160. [Google Scholar] [CrossRef]
- Cripps, D.; Thomas, S.N.; Jeng, Y.; Yang, F.; Davies, P.; Yang, A.J. Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J. Biol. Chem. 2006, 281, 10825–10838. [Google Scholar] [CrossRef] [Green Version]
- Petrucelli, L. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 2004, 13, 703–714. [Google Scholar] [CrossRef] [Green Version]
- Munari, F.; Barracchia, C.G.; Franchin, C.; Parolini, F.; Capaldi, S.; Romeo, A.; Bubacco, L.; Assfalg, M.; Arrigoni, G.; D’Onofrio, M. Semisynthetic and Enzyme-Mediated Conjugate Preparations Illuminate the Ubiquitination-Dependent Aggregation of Tau Protein. Angew. Chem. Int. Ed. 2020. [Google Scholar] [CrossRef]
- Mali, S.M.; Singh, S.K.; Eid, E.; Brik, A. Ubiquitin Signaling: Chemistry Comes to the Rescue. J. Am. Chem. Soc. 2017, 139, 4971–4986. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Abeywardana, T.; Dhall, A.; Marotta, N.P.; Varkey, J.; Langen, R.; Chatterjee, C.; Pratt, M.R. Semisynthetic, Site-Specific Ubiquitin Modification of α-Synuclein Reveals Differential Effects on Aggregation. J. Am. Chem. Soc. 2012, 134, 5468–5471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Ai, Y.; Wang, J.; Haracska, L.; Zhuang, Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat. Chem. Biol. 2010, 6, 270–272. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, C.; McGinty, R.K.; Fierz, B.; Muir, T.W. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat. Chem. Biol. 2010, 6, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Barghorn, S.; Biernat, J.; Mandelkow, E. Purification of Recombinant Tau Protein and Preparation of Alzheimer-Paired Helical Filaments In Vitro. In Amyloid Proteins; Humana Press: Totowa, NJ, USA, 2004; Volume 299, pp. 035–052. ISBN 978-1-59259-874-8. [Google Scholar]
- Kayed, R. Common Structure of Soluble Amyloid Oligomers Implies Common Mechanism of Pathogenesis. Science 2003, 300, 486–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haj-Yahya, M.; Gopinath, P.; Rajasekhar, K.; Mirbaha, H.; Diamond, M.I.; Lashuel, H.A. Site-Specific Hyperphosphorylation Inhibits, Rather than Promotes, Tau Fibrillization, Seeding Capacity, and Its Microtubule Binding. Angew. Chem. Int. Ed. 2020, 59, 4059–4067. [Google Scholar] [CrossRef] [Green Version]
- Livneh, I.; Kravtsova-Ivantsiv, Y.; Braten, O.; Kwon, Y.T.; Ciechanover, A. Monoubiquitination joins polyubiquitination as an esteemed proteasomal targeting signal. BioEssays 2017, 39, 1700027. [Google Scholar] [CrossRef]
- Arosio, P.; Knowles, T.P.J.; Linse, S. On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 2015, 17, 7606–7618. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, D.; Walinda, E.; Fukada, H.; Sou, Y.-S.; Kageyama, S.; Hoshino, M.; Fujii, T.; Tsuchiya, H.; Saeki, Y.; Arita, K.; et al. The unexpected role of polyubiquitin chains in the formation of fibrillar aggregates. Nat. Commun. 2015, 6, 6116. [Google Scholar] [CrossRef] [Green Version]
- Ellmer, D.; Brehs, M.; Haj-Yahya, M.; Lashuel, H.A.; Becker, C.F.W. Single Posttranslational Modifications in the Central Repeat Domains of Tau4 Impact its Aggregation and Tubulin Binding. Angew. Chem. Int. Ed. 2019, 58, 1616–1620. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Falcon, B.; Murzin, A.G.; Fan, J.; Crowther, R.A.; Goedert, M.; Scheres, S.H. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. eLife 2019, 8, e43584. [Google Scholar] [CrossRef] [PubMed]
- Flach, K.; Ramminger, E.; Hilbrich, I.; Arsalan-Werner, A.; Albrecht, F.; Herrmann, L.; Goedert, M.; Arendt, T.; Holzer, M. Axotrophin/MARCH7 acts as an E3 ubiquitin ligase and ubiquitinates tau protein in vitro impairing microtubule binding. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2014, 1842, 1527–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munari, F.; Bortot, A.; Zanzoni, S.; D’Onofrio, M.; Fushman, D.; Assfalg, M. Identification of primary and secondary UBA footprints on the surface of ubiquitin in cell-mimicking crowded solution. FEBS Lett. 2017, 591, 979–990. [Google Scholar] [CrossRef] [Green Version]
- Munari, F.; Bortot, A.; Assfalg, M.; D’Onofrio, M. Alzheimer’s disease-associated ubiquitin mutant Ubb+1: Properties of the carboxy-terminal domain and its influence on biomolecular interactions. Int. J. Biol. Macromol. 2018, 108, 24–31. [Google Scholar] [CrossRef]
- Varadan, R.; Assfalg, M.; Fushman, D. Ranjani Varadan; Assfalg, M.; Fushman, D. Using NMR Spectroscopy to Monitor Ubiquitin Chain Conformation and Interactions with Ubiquitin-Binding Domains. Methods Enzymol. 2005, 399, 177–192. [Google Scholar]
- Bortot, A.; Zanzoni, S.; D’Onofrio, M.; Assfalg, M. Specific Interaction Sites Determine Differential Adsorption of Protein Structural Isomers on Nanoparticle Surfaces. Chem. Eur. J. 2018, 24, 5911–5919. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, L.; Khurana, R.; Coats, A.; Frokjaer, S.; Brange, J.; Vyas, S.; Uversky, V.N.; Fink, A.L. Effect of Environmental Factors on the Kinetics of Insulin Fibril Formation: Elucidation of the Molecular Mechanism. Biochemistry 2001, 40, 6036–6046. [Google Scholar] [CrossRef] [PubMed]






| t0.5 (h) | tlag (h) | τ (h) | |
|---|---|---|---|
| tau4RDΔC | 5.2 ± 0.1 | 3.8 ± 0.5 | 0.7 ± 0.2 |
| Ub-tau4RD(353) | 14.8 ± 1.0 | 12.4 ± 0.1 | 1.1 ± 0.6 |
| Ub2(48)-tau4RD(353) | 20.7 ± 1.2 | 12.1 ± 1.1 | 4.3 ± 0.1 |
| Ub2(63)-tau4RD(353) | 21.1 ± 3.3 | 15.1 ± 1.4 | 3.0 ± 0.9 |
| Large Width | Narrow Width | Crossover Distance | |
|---|---|---|---|
| tau4RDΔC | 22 ± 2 nm (n = 14) | 12 ± 2 nm (n = 13) | 74 ± 8 nm (n = 24) |
| Ub-tau4RD(353) | 19 ± 2 nm (n = 20) | 12 ± 2 nm (n = 22) | 56 ± 6 nm (n = 31) |
| Ub2(48)-tau4RD(353) | 20 ± 2 nm (n =19) | 16 ± 2 nm (n = 19) | 58 ± 6 nm (n = 27) |
| Ub2(63)-tau4RD(353) | 20 ± 2 nm (n = 26) | 15 ± 2 nm (n = 24) | 57 ± 8 nm (n = 37) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munari, F.; Barracchia, C.G.; Parolini, F.; Tira, R.; Bubacco, L.; Assfalg, M.; D’Onofrio, M. Semisynthetic Modification of Tau Protein with Di-Ubiquitin Chains for Aggregation Studies. Int. J. Mol. Sci. 2020, 21, 4400. https://doi.org/10.3390/ijms21124400
Munari F, Barracchia CG, Parolini F, Tira R, Bubacco L, Assfalg M, D’Onofrio M. Semisynthetic Modification of Tau Protein with Di-Ubiquitin Chains for Aggregation Studies. International Journal of Molecular Sciences. 2020; 21(12):4400. https://doi.org/10.3390/ijms21124400
Chicago/Turabian StyleMunari, Francesca, Carlo Giorgio Barracchia, Francesca Parolini, Roberto Tira, Luigi Bubacco, Michael Assfalg, and Mariapina D’Onofrio. 2020. "Semisynthetic Modification of Tau Protein with Di-Ubiquitin Chains for Aggregation Studies" International Journal of Molecular Sciences 21, no. 12: 4400. https://doi.org/10.3390/ijms21124400
APA StyleMunari, F., Barracchia, C. G., Parolini, F., Tira, R., Bubacco, L., Assfalg, M., & D’Onofrio, M. (2020). Semisynthetic Modification of Tau Protein with Di-Ubiquitin Chains for Aggregation Studies. International Journal of Molecular Sciences, 21(12), 4400. https://doi.org/10.3390/ijms21124400