An Intact Cell Bioluminescence-Based Assay for the Simple and Rapid Diagnosis of Urinary Tract Infection
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Bioluminescent Bacteria
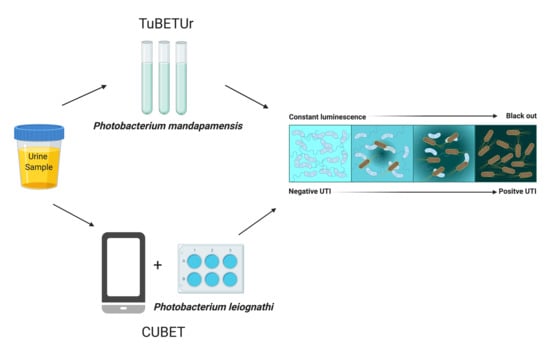
2.2. Tube Bioluminescence Extinction Technology for Urine (TuBETUr)
2.3. Relationship between the Cell Density of Selected Uropathogens and Time-To-Blackout in Artificial Urine
2.4. Application of TuBETUr for UTI Diagnosis
2.5. Optimization of Lyophilization Conditions for Photobacterium leiognathi ATCC 33981TM
2.6. Microtiter Plate Assay Using Lyophilized Photobacterium leiognathi
2.7. Mechanism of Action for Bioluminescence Inhibition
2.8. Cellphone-Based Urinary Tract Infection Bioluminescence Extinction Technology (CUBET)
3. Materials and Methods
3.1. Formulation of the Agar Medium for Bioluminescent Bacteria
3.2. Isolation of Bioluminescent Bacteria
3.3. Identification of Bioluminescent Bacteria from Squid (Phenotypic)
3.4. Identification of Bioluminescent Bacteria (Genotypic)
3.5. Preparation of Luminous Bacterial Suspension in 2.5% (w/v) Saline
3.6. Urine Samples
3.7. Preparation of Artificial Urine (AU-Siriraj)
3.8. Bioluminescence Assay Using Reference Organisms in Artificial Urine for the Tube Bioluminescence Extinction Assay Urine (TuBETUr)
3.9. Tube Bioluminescence Extinction Technology for Urine (TuBETUr) Samples
3.10. Assay to Determine the Mechanism of Action
Determination of Acyl Homoserine Lactone (AHL), Autoinducer 2 (AI2), and Bacterial Toxin Production Effects on Non-Pathogenic E. coli
3.11. Detection of UTI in Artificial Urine Using the CUBET Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| UTI | Urinary tract infection |
| TuBETUr | Tube bioluminescence extinction technology urine |
| CUBET | Cellphone-based UTI bioluminescence extinction technology |
| CFU | Colony-forming unit |
| TYESA | Trypticase yeast extract seawater agar |
| ATCC | American Type Culture Collection |
| USTCMS | University of Santo Tomas Collection of Microbial Strains |
| LE | Leukocyte esterase |
| NT | Nitrite test |
| FISH | Fluorescence in situ hybridization |
| ATP | Adenosine triphosphate |
| RNA | Ribonucleic acid |
| rRNA | Ribosomal ribonucleic acid |
| BLAST | Basic Local Alignment Search Tool |
| Bp | Base pair |
| pH | Potential of hydrogen |
| AHL | Acyl homoserine lactone |
| AI2 | Autoinducer-2 |
| GI | Gastrointestinal |
| APW | Alkaline peptone water |
| SIM | Sulfur indole motility |
| TSB | Trypticase soy broth |
| TCBS | Thiosulfate citrate bile salt sucrose |
| RLU | Relative light unit |
| OD | Optical density |
| rpm | Rotation per min |
| DSMZ | Deutsche Sammlung von Mikroorganismen und Zellkulturen |
References
- Terlizzi, M.E.; Gribaudo, G.; Maffei, M.E. UroPathogenic Escherichia coli (UPEC) infections: Virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front. Microbiol. 2017, 8, 1566. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Ejrnæs, K. Bacterial characteristics of importance for recurrent urinary tract infections caused by Escherichia coli. Dan. Med. Bull. 2011, 58, B4187. [Google Scholar] [PubMed]
- Ho, H.J.; Tan, M.X.; Chen, M.I.; Tan, T.Y.; Koo, S.H.; Koong, A.Y.; Ng, L.P.; Hu, P.L.; Tan, K.T.; Moey, P.K. Interaction between antibiotic resistance, resistance genes, and treatment response for urinary tract infections in primary care. J. Clin. Microbiol. 2019, 57, e00143-19. [Google Scholar] [CrossRef] [Green Version]
- Klein, R.D.; Hultgren, S.J. Urinary tract infections: Microbial pathogenesis, host–pathogen interactions and new treatment strategies. Nat. Rev. Microbiol. 2020, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mach, K.E.; Wong, P.K.; Liao, J.C. Biosensor diagnosis of urinary tract infections: A path to better treatment? Trends Pharmacol. Sci. 2011, 32, 330–336. [Google Scholar] [CrossRef] [Green Version]
- Males, B.M.; Bartholomew, W.R.; Amsterdam, D. Leukocyte esterase-nitrite and bioluminescence assays as urine screens. J. Clin. Microbiol. 1985, 22, 531–534. [Google Scholar] [CrossRef] [Green Version]
- Hughes, J.G.; Snyder, R.J.; Washington, J.A., II. An evaluation of a leukocyte esterase/nitrite test strip and a bioluminescence assay for detection of bacteriuria. Diagn. Microbiol. Infect. Dis. 1985, 3, 139–142. [Google Scholar] [CrossRef]
- Flenker, K.S.; Burghardt, E.L.; Dutta, N.; Burns, W.J.; Grover, J.M.; Kenkel, E.J.; Weaver, T.M.; Mills, J.; Kim, H.; Huang, L. Rapid detection of urinary tract infections via bacterial nuclease activity. Mol. Ther. 2017, 25, 1353–1362. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.T.; Heneghan, C.; Price, C.P.; Van den Bruel, A.; Plüddemann, A. Point-Of-Care Testing for Urinary Tract Infections; Horizon Scan Report 0045; NIHR: Woodstock Road Oxford, UK, 2016; Available online: https://www.community.healthcare.mic.nihr.ac.uk/reports-and-resources/horizon-scanning-reports/point-of-care-testing-for-urinary-tract-infections (accessed on 14 July 2020).
- Ivnitski, D.; Abdel-Hamid, I.; Atanasov, P.; Wilkins, E. Biosensors for detection of pathogenic bacteria. Biosens. Bioelectron. 1999, 14, 599–624. [Google Scholar] [CrossRef]
- Galloway, A.; Graham, J. The laboratory diagnosis of urinary tract infection.(ACP Best Practice No 167). J. Clin. Pathol. 2001, 54, 911–920. [Google Scholar]
- Van Nostrand, J.D.; Junkins, A.D.; Bartholdi, R.K. Poor predictive ability of urinalysis and microscopic examination to detect urinary tract infection. Am. J. Clin. Pathol. 2000, 113, 709–713. [Google Scholar] [CrossRef]
- Davenport, M.; Mach, K.E.; Shortliffe, L.M.D.; Banaei, N.; Wang, T.-H.; Liao, J.C. New and developing diagnostic technologies for urinary tract infections. Nat. Rev. Urol. 2017, 14, 296. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Chhibber, S.; Harjai, K. Quorum sensing is necessary for the virulence of Pseudomonas aeruginosa during urinary tract infection. Kidney Int. 2009, 76, 286–292. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Chhibber, S.; Gupta, V.; Harjai, K. Screening & profiling of quorum sensing signal molecules in Pseudomonas aeruginosa isolates from catheterized urinary tract infection patients. Indian J. Med. Res. 2011, 134, 208. [Google Scholar]
- Gao, Y.; Lin, Z.; Chen, R.; Wang, T.; Liu, S.; Yao, Z.; Yin, D. Using molecular docking to compare toxicity of reactive chemicals to freshwater and marine luminous bacteria. Mol. Inform. 2012, 31, 809–816. [Google Scholar] [CrossRef]
- Bastholm, S.; Wahlstrøm, L.; Bjergbæk, L.A.; Roslev, P. A simple bioluminescence procedure for early warning detection of coliform bacteria in drinking water. World J. Microbiol. Biotechnol. 2008, 24, 2323–2330. [Google Scholar] [CrossRef]
- Quinto, E.A. A simple water toxicity test using Photobcicterium leiognathi. J. Biol. Educ. 2001, 35, 89–92. [Google Scholar] [CrossRef]
- Kolbeck, J.C.; Padgett, R.; Estevez, E.G.; Harrell, L.J. Bioluminescence screening for bacteriuria. J. Clin. Microbiol. 1985, 21, 527–530. [Google Scholar] [CrossRef] [Green Version]
- Selan, L.; Berlutti, F.; Passariello, C.; Thaller, M.C.; Renzini, G. Reliability of a bioluminescence ATP assay for detection of bacteria. J. Clin. Microbiol. 1992, 30, 1739–1742. [Google Scholar] [CrossRef] [Green Version]
- Hassan, S.H.; Oh, S.E. Improved detection of toxic chemicals by Photobacterium phosphoreum using modified Boss medium. J. Photochem. Photobiol. B Biol. 2010, 101, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Tanet, L.; Tamburini, C.; Baumas, C.; Garel, M.; Simon, G.; Casalot, L. Bacterial Bioluminescence: Light Emission in Photobacterium phosphoreum Is Not Under Quorum-Sensing Control. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Ngamsom, B.; Wandera, E.A.; Iles, A.; Kimani, R.; Muregi, F.; Gitaka, J.; Pamme, N. Rapid detection of Group B Streptococcus (GBS) from artificial urine samples based on IFAST and ATP bioluminescence assay: From development to practical challenges during protocol testing in Kenya. Analyst 2019, 144, 6889–6897. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, P.V.; Ast, J.C.; Kimura, S.; Fukui, A.; Yoshino, T.; Endo, H. Phylogenetic analysis of host–symbiont specificity and codivergence in bioluminescent symbioses. Cladistics 2007, 23, 507–532. [Google Scholar] [CrossRef]
- Kaeding, A.J.; Ast, J.C.; Pearce, M.M.; Urbanczyk, H.; Kimura, S.; Endo, H.; Nakamura, M.; Dunlap, P.V. Phylogenetic diversity and cosymbiosis in the bioluminescent symbioses of “Photobacterium mandapamensis”. Appl. Environ. Microbiol. 2007, 73, 3173–3182. [Google Scholar] [CrossRef] [Green Version]
- Pezzlo, M. Detection of urinary tract infections by rapid methods. Clin. Microbiol. Rev. 1988, 1, 268–280. [Google Scholar] [CrossRef]
- Liu, B.-F.; Ozaki, M.; Hisamoto, H.; Luo, Q.; Utsumi, Y.; Hattori, T.; Terabe, S. Microfluidic chip toward cellular ATP and ATP-conjugated metabolic analysis with bioluminescence detection. Anal. Chem. 2005, 77, 573–578. [Google Scholar] [CrossRef]
- Ivančić, V.; Mastali, M.; Percy, N.; Gornbein, J.; Babbitt, J.T.; Li, Y.; Landaw, E.M.; Bruckner, D.A.; Churchill, B.M.; Haake, D.A. Rapid antimicrobial susceptibility determination of uropathogens in clinical urine specimens by use of ATP bioluminescence. J. Clin. Microbiol. 2008, 46, 1213–1219. [Google Scholar] [CrossRef] [Green Version]
- Quinto, E.A. Simple and Novel Methods of the Evaluation of Water Pollution and Water Pollutants Using Bacterial Bioluminescence in Suspensions and Immobilized Forms. Philipp. Biota Xl 2007, 1, 3–13. [Google Scholar]
- Quinto, E.A. Disc Immobilized Bioluminescence Technology (PIBIT): A Novel, Simple and Inexpensive Method of Determining the Antibacterial Activity of Antiseptics, Disinfectants and Health-Care Products. Int. J. Antimicrob. Agents 2005, 26, 109–110. [Google Scholar]
- Rowe, T.A.; Juthani-Mehta, M. Diagnosis and management of urinary tract infection in older adults. Infect. Dis. Clin. N. Am. 2014, 28, 75. [Google Scholar] [CrossRef] [Green Version]
- Leigh, D.; Williams, J. Method for the detection of significant bacteriuria in large groups of patients. J. Clin. Pathol. 1964, 17, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Grabe, M.; Bjerklund-Johansen, T.; Botto, H.; Çek, M.; Naber, K.; Tenke, P.; Wagenlehner, F. Guidelines on urological infections. Eur. Assoc. Urol. 2015, 182. [Google Scholar]
- Reichelt, J.L.; Baumann, P. Photobacterium mandapamensis Hendrie et al., a Later Subjective Synonym of Photobacterium leiognathi Boisvert et al. Int. J. Syst. Evol. Microbiol. 1975, 25, 208–209. [Google Scholar] [CrossRef] [Green Version]
- Urbanczyk, H.; Ast, J.C.; Dunlap, P.V. Phylogeny, genomics, and symbiosis of Photobacterium. FEMS Microbiol. Rev. 2011, 35, 324–342. [Google Scholar] [CrossRef] [PubMed]
- Camanzi, L.; Bolelli, L.; Maiolini, E.; Girotti, S.; Matteuzzi, D. Optimal conditions for stability of photoemission and freeze drying of two luminescent bacteria for use in a biosensor. Environ. Toxicol. Chem. 2011, 30, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [Green Version]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [Green Version]
- Dunn, A.K.; Rader, B.A.; Stabb, E.V.; Mandel, M.J. Regulation of bioluminescence in Photobacterium leiognathi strain KNH6. J. Bacteriol. 2015, 197, 3676–3685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brook, I. Urinary tract and genito-urinary suppurative infections due to anaerobic bacteria. Int. J. Urol. 2004, 11, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Riedel, T.E.; Berelson, W.M.; Nealson, K.H.; Finkel, S.E. Oxygen Consumption Rates of Bacteria under Nutrient-Limited Conditions. Appl. Environ. Microbiol. 2013, 79, 4921. [Google Scholar] [CrossRef] [Green Version]
- Fukasawa, S.; Dunlap, P.V. Identification of luminous bacteria isolated from the light organ of the squid, Doryteuthis kensaki. Agric. Biol. Chem. 1986, 50, 1645–1646. [Google Scholar]
- Malave-Orengo, J.; Rubio-Marrero, E.N.; Rios-Velazquez, C. Isolation and characterization of bioluminescent bacteria from marine environments of Puerto Rico. Cur. Res. Technol. Educ. Top. Appl. Microbiol. Microb. Biotechnol. 2010, 1, 103–108. Available online: https://pdfs.semanticscholar.org/734f/3a7b9c79768bc8e39d74449c0e0317018561.pdf (accessed on 15 July 2020).
- Chutipongtanate, S.; Thongboonkerd, V. Systematic comparisons of artificial urine formulas for in vitro cellular study. Anal. Biochem. 2010, 402, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]





| UROPATHOGEN | Time of Blackout (s) | Regression Equation (R2) | 105 cfu/mL (s) * | |
|---|---|---|---|---|
| 102 cfu/mL | 108 cfu/mL | |||
| Escherichia coli ATCC 25922TM | 1242 | 316 | 0.9859 | 831 |
| Staphylococcus aureus ATCC 23235TM | 1145 | 253 | 0.9953 | 663 |
| Proteus mirabilis ATCC 35659TM | 1431 | 238 | 0.9842 | 694 |
| Candida albicans ATCC 14053TM | 1978 | 392 | 0.9851 | 1246 |
| Cut-off time | - | 1257 s | - | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyes, S.; Le, N.; Fuentes, M.D.; Upegui, J.; Dikici, E.; Broyles, D.; Quinto, E.; Daunert, S.; Deo, S.K. An Intact Cell Bioluminescence-Based Assay for the Simple and Rapid Diagnosis of Urinary Tract Infection. Int. J. Mol. Sci. 2020, 21, 5015. https://doi.org/10.3390/ijms21145015
Reyes S, Le N, Fuentes MD, Upegui J, Dikici E, Broyles D, Quinto E, Daunert S, Deo SK. An Intact Cell Bioluminescence-Based Assay for the Simple and Rapid Diagnosis of Urinary Tract Infection. International Journal of Molecular Sciences. 2020; 21(14):5015. https://doi.org/10.3390/ijms21145015
Chicago/Turabian StyleReyes, Sherwin, Nga Le, Mary Denneth Fuentes, Jonathan Upegui, Emre Dikici, David Broyles, Edward Quinto, Sylvia Daunert, and Sapna K. Deo. 2020. "An Intact Cell Bioluminescence-Based Assay for the Simple and Rapid Diagnosis of Urinary Tract Infection" International Journal of Molecular Sciences 21, no. 14: 5015. https://doi.org/10.3390/ijms21145015
APA StyleReyes, S., Le, N., Fuentes, M. D., Upegui, J., Dikici, E., Broyles, D., Quinto, E., Daunert, S., & Deo, S. K. (2020). An Intact Cell Bioluminescence-Based Assay for the Simple and Rapid Diagnosis of Urinary Tract Infection. International Journal of Molecular Sciences, 21(14), 5015. https://doi.org/10.3390/ijms21145015