Extracellular Nucleotides Selectively Induce Migration of Chondrocytes and Expression of Type II Collagen
Abstract
:1. Introduction
2. Results
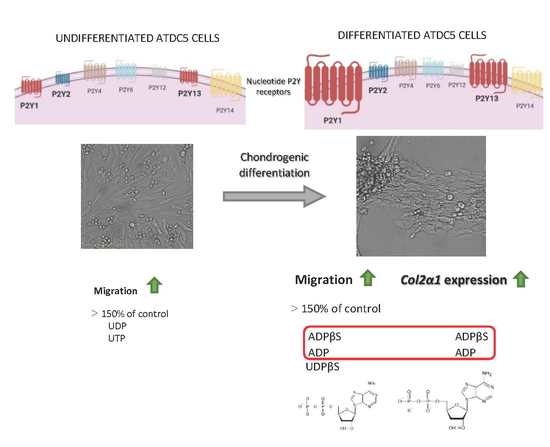
2.1. The Chondrogenic Phenotype of ATDC5 Growing Under Differentiating Conditions
2.2. Expression of P2Y Receptors in Differentiated and Undifferentiated ATDC5 Cells
2.3. Migration of ATDC5 Cells in the Presence of Extracellular Nucleotides
2.4. The Influence of Extracellular Nucleotides on the Expression of Cartilage-Related Gene Markers in Differentiated and Undifferentiated ATDC5 Cells
3. Discussion
4. Materials and Methods
4.1. Tested Compounds
4.2. ATDC5 Cell Culture
4.3. Culture Media and Cell Differentiation
4.4. RNA Isolation and Reverse Transcription Quantitative Polymerase Chain Reaction
4.5. Cell Migration Studies
4.6. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACI | Autologous chondrocyte implantation |
| ADP | Adenosine 5′-O-diphosphate |
| ADPβS | Adenosine 5′-O-β-thiodiphosphate |
| AMPS | Adenosine 5′-O-thiomonophosphate |
| ATP | Adenosine 5′-O-triphosphate |
| ATPγS | Adenosine 5′-O-γ-thiotriphosphate |
| DMEM | Dulbecco’s modified Eagle medium |
| FBS | Fetal Bovine Serum |
| ITS | Insulin-Transferrin-Sodium Selenite |
| MSCs | Mesenchymal stem cells |
| OA | Osteoarthritis |
| PBS | Phosphate-buffered saline |
| RT-qPCR | Reverse transcription quantitative polymerase chain reaction |
| SEM | Standard error of the mean |
| TGF-β2 | Transforming Growth Factor β2 |
| TMPS | Thymidine 5′-O-thiomonophosphate |
| UDP | Uridine 5′-O-diphosphate |
| UDPβS | Uridine 5′-O-β-thiodiphosphate |
| UMPS | Uridine 5′-O-thiomonophosphate |
| UTP | Uridine 5′-O-triphosphate |
| UTPγS | Uridine 5′-O-γ-thiotriphosphate |
References
- Akkiraju, H.; Nohe, A. Role of chondrocytes in cartilage formation, progression of osteoarthritis and cartilage regeneration. J. Dev. Biol. 2015, 3, 177–192. [Google Scholar] [CrossRef] [Green Version]
- Hopper, N.; Henson, F.; Brooks, R.; Ali, E.; Rushton, N.; Wardale, J. Peripheral blood derived mononuclear cells enhance osteoarthritic human chondrocyte migration. Arthritis Res. Ther. 2015, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.; Lauffenburger, D.A.; Morales, T.I. Motile chondrocytes from newborn calf: Migration properties and synthesis of collagen II. Osteoarthr. Cartil. 2003, 11, 603–612. [Google Scholar] [CrossRef] [Green Version]
- Maniwa, S.; Ochi, M.; Motomura, T.; Nishikori, T.; Chen, J.; Naora, H. Effects of hyaluronic acid and basic fibroblast growth factor on motility of chondrocytes and synovial cells in culture. Acta Orthop. Scand. 2001, 72, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, D.W.; Riehle, M.O.; Rappuoli, R.; Monaghan, W.; Barbucci, R.; Curtis, A.S.G. The response of primary articular chondrocytes to micrometric surface topography and sulphated hyaluronic acid-based matrices. Cell Biol. Int. 2005, 29, 605–615. [Google Scholar] [CrossRef]
- Kirilak, Y.; Pavlos, N.J.; Willers, C.R.; Han, R.; Feng, H.; Xu, J.; Asokananthan, N.; Stewart, G.A.; Henry, P.; Wood, D.; et al. Fibrin sealant promotes migration and proliferation of human articular chondrocytes: Possible involvement of thrombin and protease-activated receptors. Int. J. Mol. Med. 2006, 17, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Shi, R.; Ma, X. The secreted protein WNT5A regulates condylar chondrocyte proliferation, hypertrophy and migration. Arch. Oral Biol. 2017, 82, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Węgłowska, E.; Szustak, M.; Gendaszewska-Darmach, E. Proangiogenic properties of nucleoside 5′-O-phosphorothioate analogues under hyperglycaemic conditions. Curr. Top. Med. Chem. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Gendaszewska-Darmach, E.; Szustak, M. Thymidine 5′-O-monophosphorothioate induces HeLa cell migration by activation of the P2Y6 receptor. Purinergic Signal. 2016, 12, 199–209. [Google Scholar] [CrossRef] [Green Version]
- Jacobson, K.A.; Delicado, E.G.; Gachet, C.; Kennedy, C.; Kügelgen, I.; Li, B.; Miras-Portugal, M.T.; Novak, I.; Schöneberg, T.; Perez-Sen, R.; et al. Update of P2Y receptor pharmacology: IUPHAR Review 27. Br. J. Pharmacol. 2020, 177, 2413–2433. [Google Scholar] [CrossRef]
- Guzmán-Aránguez, A.; Irazu, M.; Yayon, A.; Pintor, J. P2Y receptors activated by diadenosine polyphosphates reestablish Ca2+ transients in achondroplasic chondrocytes. Bone 2008, 42, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Caswell, A.M.; Russell, R.G.G. Identification of ecto-nucleoside triphosphate pyrophosphatase in human articular chondrocytes in monolayer culture. BBA Mol. Cell Res. 1985, 847, 40–47. [Google Scholar] [CrossRef]
- Drzazga, A.; Sowinska, A.; Krzeminska, A.; Rytczak, P.; Koziolkiewicz, M.; Gendaszewska-Darmach, E. Lysophosphatidylcholine elicits intracellular calcium signaling in a GPR55-dependent manner. Biochem. Biophys. Res. Commun. 2017, 489. [Google Scholar] [CrossRef] [PubMed]
- Haanes, K.A.; Edvinsson, L. Expression and characterization of purinergic receptors in rat middle meningeal artery-potential role in migraine. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Quintas, C.; Vale, N.; Gonçalves, J.; Queiroz, G. Microglia P2Y13 receptors prevent astrocyte proliferation mediated by P2Y1 receptors. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Davies, R.L.; Kuiper, N.J. Regenerative Medicine: A Review of the Evolution of Autologous Chondrocyte Implantation (ACI) Therapy. Bioengineering 2019, 6, 22. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.F.; Samsa, W.E.; Zhou, G.; Lefebvre, V. Transcriptional control of chondrocyte specification and differentiation. Semin. Cell Dev. Biol. 2017, 62, 34–49. [Google Scholar] [CrossRef] [Green Version]
- Ushijima, T.; Okazaki, K.; Tsushima, H.; Iwamoto, Y. CCAAT/enhancer-binding protein β regulates the repression of type II collagen expression during the differentiation from proliferative to hypertrophic chondrocytes. J. Biol. Chem. 2014, 289, 2852–2863. [Google Scholar] [CrossRef] [Green Version]
- Caron, M.M.J.; Emans, P.J.; Coolsen, M.M.E.; Voss, L.; Surtel, D.A.M.; Cremers, A.; van Rhijn, L.W.; Welting, T.J.M. Redifferentiation of dedifferentiated human articular chondrocytes: Comparison of 2D and 3D cultures. Osteoarthr. Cartil. 2012, 20, 1170–1178. [Google Scholar] [CrossRef] [Green Version]
- Challa, T.D.; Rais, Y.; Ornan, E.M. Effect of adiponectin on ATDC5 proliferation, differentiation and signaling pathways. Mol. Cell. Endocrinol. 2010, 323, 282–291. [Google Scholar] [CrossRef]
- Ustun, S.; Tombuloglu, A.; Kilinc, M.; Guler, M.O.; Tekinay, A.B. Growth and differentiation of prechondrogenic cells on bioactive self-assembled peptide nanofibers. Biomacromolecules 2013, 14, 17–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Zhai, Z.; Wang, Y. Evaluation of insulin medium or chondrogenic medium on proliferation and chondrogenesis of ATDC5 cells. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Barlic, A.; Radosavljevic, D.; Drobnic, M.; Kregar-Velikonja, N. Advancing in the quality of the cells assigned for autologous chondrocyte implantation (ACI) method. In IFMBE Proceedings; Springer: Berlin/Heidelberg, Germany, 2007; Volume 16, pp. 249–252. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dreisig, K.; Kornum, B.R. A critical look at the function of the P2Y11 receptor. Purinergic Signal. 2016, 12, 427–437. [Google Scholar] [CrossRef] [Green Version]
- Morales, T.I. Chondrocyte moves: Clever strategies? Osteoarthr. Cartil. 2007, 15, 861–871. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Tuan, R.S. Origin and function of cartilage stem/progenitor cells in osteoarthritis. Nat. Rev. Rheumatol. 2015, 11, 206–212. [Google Scholar] [CrossRef] [Green Version]
- Schubert, T.; Anders, S.; Neumann, E.; Schölmerich, J.; Hofstädter, F.; Grifka, J.; Müller-Ladner, U.; Libera, J.; Schedel, J. Long-term effects of chondrospheres on cartilage lesions in an autologous chondrocyte implantation model as investigated in the SCID mouse model. Int. J. Mol. Med. 2009, 23, 455–460. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Xu, Y.; Yin, Z.; Yang, X.; Jiang, Y.; Gui, J. Chondrocyte migration affects tissue-engineered cartilage integration by activating the signal transduction pathways involving Src, PLCγ1, and ERK1/2. Tissue Eng. Part A 2013, 19, 2506–2516. [Google Scholar] [CrossRef]
- Matta, C.; Fodor, J.; Csernoch, L.; Zákány, R. Purinergic signalling-evoked intracellular Ca2+ concentration changes in the regulation of chondrogenesis and skeletal muscle formation. Cell Calcium 2016, 59, 108–116. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Gu, Y.; Gomes, R.R.; You, J. P2Y2 receptors and GRK2 are involved in oscillatory fluid flow induced ERK1/2 responses in chondrocytes. J. Orthop. Res. 2011, 29, 828–833. [Google Scholar] [CrossRef] [Green Version]
- Kenichi, N.; Tatsumi, M.; Zhao, M. An essential and synergistic role of purinergic signaling in guided migration of corneal epithelial cells in physiological electric fields. Cell. Physiol. Biochem. 2019, 52, 198–211. [Google Scholar] [CrossRef]
- Girard, M.; Dagenais Bellefeuille, S.; Eiselt, É.; Brouillette, R.; Placet, M.; Arguin, G.; Longpré, J.; Sarret, P.; Gendron, F. The P2Y6 receptor signals through Gα q /Ca2+ /PKCα and Gα 13 /ROCK pathways to drive the formation of membrane protrusions and dictate cell migration. J. Cell. Physiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Corciulo, C.; Cronstein, B.N. Signaling of the purinergic system in the joint. Front. Pharmacol. 2020, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, L.M.; Rachow, J.W.; McCarty, D.J. Synovial fluid ATP: A potential substrate for the production of inorganic pyrophosphate. J. Rheumatol. 1991, 18, 716–720. [Google Scholar] [PubMed]
- Lazarowski, E.R.; Harden, T.K. Quantitation of extracellular UTP using a sensitive enzymatic assay. Br. J. Pharmacol. 1999, 127, 1272–1278. [Google Scholar] [CrossRef] [Green Version]
- Garcia, M.; Knight, M.M. Cyclic loading opens hemichannels to release ATP as part of a chondrocyte mechanotransduction pathway. J. Orthop. Res. 2010, 28, 510–515. [Google Scholar] [CrossRef]
- Millward-Sadler, S.J.; Wright, M.O.; Flatman, P.W.; Salter, D.M. ATP in the mechanotransduction pathway of normal human chondrocytes. Biorheology 2004, 41, 567–575. [Google Scholar]
- Knight, M.M.; McGlashan, S.R.; Garcia, M.; Jensen, C.G.; Poole, C.A. Articular chondrocytes express connexin 43 hemichannels and P2 receptors—A putative mechanoreceptor complex involving the primary cilium? J. Anat. 2009, 214, 275–283. [Google Scholar] [CrossRef]
- Koolpe, M.; Pearson, D.; Benton, H.P. Expression of both P1 and P2 purine receptor genes by human articular chondrocytes and profile of ligand-mediated prostaglandin E2 release. Arthritis Rheum. 1999, 42, 258–267. [Google Scholar] [CrossRef]
- Ciciarello, M.; Zini, R.; Rossi, L.; Salvestrini, V.; Ferrari, D.; Manfredini, R.; Lemoli, R.M. Extracellular purines promote the differentiation of human bone marrow-derived mesenchymal stem cells to the osteogenic and adipogenic lineages. Stem Cells Dev. 2013, 22, 1097–1111. [Google Scholar] [CrossRef] [Green Version]
- Biver, G.; Wang, N.; Gartland, A.; Orriss, I.; Arnett, T.R.; Boeynaems, J.M.; Robaye, B. Role of the P2Y13 receptor in the differentiation of bone marrow stromal cells into osteoblasts and adipocytes. Stem Cells 2013, 31, 2747–2758. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Xu, C.; Cheng, X.; Liu, Y.; Yue, M.; Hu, M.; Luo, D.; Niu, Y.; Ouyang, H.; Ji, J.; et al. Platelets promote cartilage repair and chondrocyte proliferation via ADP in a rodent model of osteoarthritis. Platelets 2016, 27, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Semenova, S.; Shatrova, A.; Vassilieva, I.; Shamatova, M.; Pugovkina, N.; Negulyaev, Y. Adenosine-5′-triphosphate suppresses proliferation and migration capacity of human endometrial stem cells. J. Cell. Mol. Med. 2020, 24, 4580–4588. [Google Scholar] [CrossRef] [PubMed]
- Leong, W.S.; Russell, R.G.G.; Caswell, A.M. Stimulation of cartilage resorption by extracellular ATP acting at P2-purinoceptors. BBA Gen. Subj. 1994, 1201, 298–304. [Google Scholar] [CrossRef]
- Brown, C.J.; Caswell, A.M.; Rahman, S.; Russell, R.G.G.; Buttle, D.J. Proteoglycan breakdown from bovine nasal cartilage is increased, and from articular cartilage is decreased, by extracellular ATP. Biochim. Biophys. Acta Mol. Basis Dis. 1997, 1362, 208–220. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, H.; Zebisch, M.; Sträter, N. Cellular function and molecular structure of ecto-nucleotidases. Purinergic Signal. 2012, 8, 437–502. [Google Scholar] [CrossRef] [Green Version]
- Graff, R.D.; Picher, M.; Lee, G.M. Extracellular nucleotides, cartilage stress, and calcium crystal formation. Curr. Opin. Rheumatol. 2003, 15, 315–320. [Google Scholar] [CrossRef]
- Kazemnejad, S.; Khanmohammadi, M.; Baheiraei, N.; Arasteh, S. Current State of Cartilage Tissue Engineering using Nanofibrous Scaffolds and Stem Cells. Avicenna J. Med. Biotechnol. 2017, 9, 50–65. [Google Scholar]




| Gene | NCBI Reference Sequence | Forward Primer | Reverse Primer |
|---|---|---|---|
| P2y1 | NM_008772 | TTATGTCAGCGTGCTGGTGT | CGTGTCTCCATTCTGCTTGA |
| P2y2 | NM_008773 | GAGGACTTCAAGTACGTGCT | ACGGAGCTGTAAGCCACAAA |
| P2y4 | NM_020621 | GGGGACAAGTATCGAAACCA | CACCCTCATAAGCAGGGAAG |
| P2y6 | NM_183168 | TGCTGTGTCAGAGGGAGTTTT | TCAGCCTTTCCTATGCTCGG |
| P2y12 | NM_001357008 | CTGAAGACCACCAGGCCATT | GGAATCCGTGCAAAGTGGAA |
| P2y13 | NM_028808 | GCACCAGAAGAGAGGCACAT | TAGGGGAAGAGTCGTCGTGT |
| P2y14 | NM_001287124 | GGAACACCCTGATCACAAAG | TGACCTTCCGTCTGACTCTT |
| Col1a1 | NM_007742 | ATGCCGCGACCTCAAGATG | TGAGGCACAGACGGCTGAGTA |
| Col2a1 | NM_031163 | AGGGCAACAGCAGGTTCACATAC | TGTCCACACCAAATTCCTGTTCA |
| Runx2 | NM_009820 | CACTGGCGGTGCAACAAGA | TTTCATAACAGCGGAGGCATTTC |
| Sox9 | NM_011448 | TCCCCGCAACAGATCTCCTA | AGGTGGAGTAGAGCCCTGAG |
| Gapdh | NM_008084 | TCTCTGCTCCTCCCTGTTCC | CAATCTCCACTTTGCCACTGC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szustak, M.; Gendaszewska-Darmach, E. Extracellular Nucleotides Selectively Induce Migration of Chondrocytes and Expression of Type II Collagen. Int. J. Mol. Sci. 2020, 21, 5227. https://doi.org/10.3390/ijms21155227
Szustak M, Gendaszewska-Darmach E. Extracellular Nucleotides Selectively Induce Migration of Chondrocytes and Expression of Type II Collagen. International Journal of Molecular Sciences. 2020; 21(15):5227. https://doi.org/10.3390/ijms21155227
Chicago/Turabian StyleSzustak, Marcin, and Edyta Gendaszewska-Darmach. 2020. "Extracellular Nucleotides Selectively Induce Migration of Chondrocytes and Expression of Type II Collagen" International Journal of Molecular Sciences 21, no. 15: 5227. https://doi.org/10.3390/ijms21155227
APA StyleSzustak, M., & Gendaszewska-Darmach, E. (2020). Extracellular Nucleotides Selectively Induce Migration of Chondrocytes and Expression of Type II Collagen. International Journal of Molecular Sciences, 21(15), 5227. https://doi.org/10.3390/ijms21155227