Long Non-Coding RNAs in Atrial Fibrillation: Pluripotent Stem Cell-Derived Cardiomyocytes as a Model System
Abstract
:1. Introduction
2. Pathophysiological Remodeling of Atria
2.1. Electrical Remodeling
2.2. Structural Remodeling
2.3. Contractile Remodeling
3. lncRNAs and Their Interventions in Heart Disease
3.1. Features and Mechanisms of lncRNAs
3.2. Functional Roles of LncRNAs in the Heart
3.2.1. LncRNAs in the Developing Heart
3.2.2. LncRNA in the Failing Heart
Cardiac Hypertrophy
Myocardial Infarction
Cardiac Fibrosis
Cardiac Arrhythmias
4. LncRNAs in Atrial Remodeling and Development of AF
4.1. LncRNAs in the Development of AF
4.2. LncRNAs in Atrial Structural Remodeling
4.3. LncRNAs in Atrial Electrical Remodeling
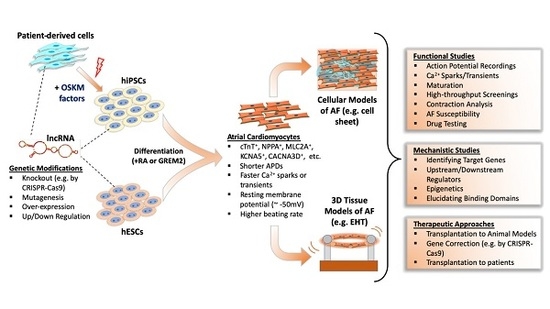
5. Deciphering LncRNA Function in Atrial Fibrillation by hPSC Disease Modeling
5.1. Differentiation and Characterization of hPSC-Derived Atrial Cardiomyocytes
5.2. Disease Modeling of AF Using hPSCs
5.3. Use of hPSCs for the Study of LncRNAs in AF
6. Challenges of PSC Modeling and Translational Aspects
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kirchhof, P.; Benussi, S.; Kotecha, D.; Ahlsson, A.; Atar, D.; Casadei, B.; Castella, M.; Diener, H.C.; Heidbuchel, H.; Hendriks, J.; et al. 2016 ESC Guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Eur. Heart J. 2016, 37, 2893–2962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conen, D.; Chae, C.U.; Glynn, R.J.; Tedrow, U.B.; Everett, B.M.; Buring, J.E.; Albert, C.M. Risk of death and cardiovascular events in initially healthy women with new-onset atrial fibrillation. JAMA 2011, 305, 2080–2087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santulli, G.; D’Ascia, S.L.; D’Ascia, C. Development of atrial fibrillation in recipients of cardiac resynchronization therapy: The role of atrial reverse remodelling. Can. J. Cardiol. 2012, 28, 245. [Google Scholar] [CrossRef] [PubMed]
- Brundel, B.J.; Van Gelder, I.C.; Henning, R.H.; Tuinenburg, A.E.; Wietses, M.; Grandjean, J.G.; Wilde, A.A.; Van Gilst, W.H.; Crijns, H.J. Alterations in potassium channel gene expression in atria of patients with persistent and paroxysmal atrial fibrillation: Differential regulation of protein and mRNA levels for K+ channels. J. Am. Coll. Cardiol. 2001, 37, 926–932. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.; Santulli, G.; Guo, X.; Gao, M.; Chen, B.X.; Marks, A.R. Imaging atrial arrhythmic intracellular calcium in intact heart. J. Mol. Cell. Cardiol. 2013, 64, 120–123. [Google Scholar] [CrossRef] [Green Version]
- Kapur, S.; Macrae, C.A. The developmental basis of adult arrhythmia: Atrial fibrillation as a paradigm. Front. Physiol. 2013, 4, 221. [Google Scholar] [CrossRef] [Green Version]
- D’Ascia, S.L.; D’Ascia, C.; Marino, V.; Lombardi, A.; Santulli, R.; Chiariello, M.; Santulli, G. Cardiac resynchronisation therapy response predicts occurrence of atrial fibrillation in non-ischaemic dilated cardiomyopathy. Int. J. Clin. Pract. 2011, 65, 1149–1155. [Google Scholar] [CrossRef]
- Shi, K.H.; Tao, H.; Yang, J.J.; Wu, J.X.; Xu, S.S.; Zhan, H.Y. Role of microRNAs in atrial fibrillation: New insights and perspectives. Cell. Signal. 2013, 25, 2079–2084. [Google Scholar] [CrossRef]
- Babapoor-Farrokhran, S.; Gill, D.; Rasekhi, R.T. The role of long noncoding RNAs in atrial fibrillation. Heart Rhythm. 2020, 17, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Dawson, K.; Wakili, R.; Ordog, B.; Clauss, S.; Chen, Y.; Iwasaki, Y.; Voigt, N.; Qi, X.Y.; Sinner, M.F.; Dobrev, D.; et al. MicroRNA29: A mechanistic contributor and potential biomarker in atrial fibrillation. Circulation 2013, 127, 1466–1475. [Google Scholar] [CrossRef] [Green Version]
- McManus, D.D.; Lin, H.; Tanriverdi, K.; Quercio, M.; Yin, X.; Larson, M.G.; Ellinor, P.T.; Levy, D.; Freedman, J.E.; Benjamin, E.J. Relations between circulating microRNAs and atrial fibrillation: Data from the Framingham Offspring Study. Heart Rhythm. 2014, 11, 663–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.; Li, L.; Zhao, S.; Yue, Y.; Yang, S. The long noncoding RNA expression profiles of paroxysmal atrial fibrillation identified by microarray analysis. Gene 2018, 642, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huang, R.; Gu, J.; Jiang, W. Identification of long non-coding RNAs as novel biomarker and potential therapeutic target for atrial fibrillation in old adults. Oncotarget 2016, 7, 10803–10811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobuss, L.; Bar, C.; Thum, T. Long Non-coding RNAs: At the Heart of Cardiac Dysfunction? Front. Physiol. 2019, 10, 30. [Google Scholar] [CrossRef] [Green Version]
- Tucker, N.R.; Clauss, S.; Ellinor, P.T. Common variation in atrial fibrillation: Navigating the path from genetic association to mechanism. Cardiovasc. Res. 2016, 109, 493–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nattel, S.; Burstein, B.; Dobrev, D. Atrial remodeling and atrial fibrillation: Mechanisms and implications. Circ. Arrhythm. Electrophysiol. 2008, 1, 62–73. [Google Scholar] [CrossRef] [Green Version]
- Schotten, U.; Verheule, S.; Kirchhof, P.; Goette, A. Pathophysiological mechanisms of atrial fibrillation: A translational appraisal. Physiol. Rev. 2011, 91, 265–325. [Google Scholar] [CrossRef]
- Shiroshita-Takeshita, A.; Mitamura, H.; Ogawa, S.; Nattel, S. Rate-dependence of atrial tachycardia effects on atrial refractoriness and atrial fibrillation maintenance. Cardiovasc. Res. 2009, 81, 90–97. [Google Scholar] [CrossRef] [Green Version]
- Allessie, M.A.; Boyden, P.A.; Camm, A.J.; Kleber, A.G.; Lab, M.J.; Legato, M.J.; Rosen, M.R.; Schwartz, P.J.; Spooner, P.M.; Van Wagoner, D.R.; et al. Pathophysiology and prevention of atrial fibrillation. Circulation 2001, 103, 769–777. [Google Scholar] [CrossRef] [Green Version]
- Wakili, R.; Voigt, N.; Kaab, S.; Dobrev, D.; Nattel, S. Recent advances in the molecular pathophysiology of atrial fibrillation. J. Clin. Invest. 2011, 121, 2955–2968. [Google Scholar] [CrossRef] [Green Version]
- Pandit, S.V.; Berenfeld, O.; Anumonwo, J.M.; Zaritski, R.M.; Kneller, J.; Nattel, S.; Jalife, J. Ionic determinants of functional reentry in a 2-D model of human atrial cells during simulated chronic atrial fibrillation. Biophys. J. 2005, 88, 3806–3821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwasaki, Y.K.; Nishida, K.; Kato, T.; Nattel, S. Atrial fibrillation pathophysiology: Implications for management. Circulation 2011, 124, 2264–2274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burstein, B.; Nattel, S. Atrial fibrosis: Mechanisms and clinical relevance in atrial fibrillation. J. Am. Coll. Cardiol. 2008, 51, 802–809. [Google Scholar] [CrossRef] [Green Version]
- Burstein, B.; Comtois, P.; Michael, G.; Nishida, K.; Villeneuve, L.; Yeh, Y.H.; Nattel, S. Changes in connexin expression and the atrial fibrillation substrate in congestive heart failure. Circ. Res. 2009, 105, 1213–1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, L.; Xie, J.; Nattel, S. Molecular determinants of cardiac fibroblast electrical function and therapeutic implications for atrial fibrillation. Cardiovasc. Res. 2011, 89, 744–753. [Google Scholar] [CrossRef] [Green Version]
- Akoum, N.; Daccarett, M.; McGann, C.; Segerson, N.; Vergara, G.; Kuppahally, S.; Badger, T.; Burgon, N.; Haslam, T.; Kholmovski, E.; et al. Atrial fibrosis helps select the appropriate patient and strategy in catheter ablation of atrial fibrillation: A DE-MRI guided approach. J. Cardiovasc. Electrophysiol. 2011, 22, 16–22. [Google Scholar] [CrossRef]
- Burstein, B.; Qi, X.Y.; Yeh, Y.H.; Calderone, A.; Nattel, S. Atrial cardiomyocyte tachycardia alters cardiac fibroblast function: A novel consideration in atrial remodeling. Cardiovasc. Res. 2007, 76, 442–452. [Google Scholar] [CrossRef] [Green Version]
- Logan, W.F.; Rowlands, D.J.; Howitt, G.; Holmes, A.M. Left Atrial Activity Following Cardioversion. Lancet 1965, 2, 471–473. [Google Scholar] [CrossRef]
- Eiras, S.; Narolska, N.A.; van Loon, R.B.; Boontje, N.M.; Zaremba, R.; Jimenez, C.R.; Visser, F.C.; Stooker, W.; van der Velden, J.; Stienen, G.J. Alterations in contractile protein composition and function in human atrial dilatation and atrial fibrillation. J. Mol. Cell. Cardiol. 2006, 41, 467–477. [Google Scholar] [CrossRef]
- Narolska, N.A.; van Loon, R.B.; Boontje, N.M.; Zaremba, R.; Penas, S.E.; Russell, J.; Spiegelenberg, S.R.; Huybregts, M.A.; Visser, F.C.; de Jong, J.W.; et al. Myocardial contraction is 5-fold more economical in ventricular than in atrial human tissue. Cardiovasc. Res. 2005, 65, 221–229. [Google Scholar] [CrossRef]
- Narolska, N.A.; Eiras, S.; van Loon, R.B.; Boontje, N.M.; Zaremba, R.; Spiegelen Berg, S.R.; Stooker, W.; Huybregts, M.A.; Visser, F.C.; van der Velden, J.; et al. Myosin heavy chain composition and the economy of contraction in healthy and diseased human myocardium. J. Muscle Res. Cell Motil. 2005, 26, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Mihm, M.J.; Yu, F.; Carnes, C.A.; Reiser, P.J.; McCarthy, P.M.; Van Wagoner, D.R.; Bauer, J.A. Impaired myofibrillar energetics and oxidative injury during human atrial fibrillation. Circulation 2001, 104, 174–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Gascoigne, D.K.; Cheetham, S.W.; Cattenoz, P.B.; Clark, M.B.; Amaral, P.P.; Taft, R.J.; Wilhelm, D.; Dinger, M.E.; Mattick, J.S. Pinstripe: A suite of programs for integrating transcriptomic and proteomic datasets identifies novel proteins and improves differentiation of protein-coding and non-coding genes. Bioinformatics 2012, 28, 3042–3050. [Google Scholar] [CrossRef]
- Banfai, B.; Jia, H.; Khatun, J.; Wood, E.; Risk, B.; Gundling, W.E., Jr.; Kundaje, A.; Gunawardena, H.P.; Yu, Y.; Xie, L.; et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012, 22, 1646–1657. [Google Scholar] [CrossRef] [Green Version]
- Dinger, M.E.; Gascoigne, D.K.; Mattick, J.S. The evolution of RNAs with multiple functions. Biochimie 2011, 93, 2013–2018. [Google Scholar] [CrossRef] [Green Version]
- Ingolia, N.T.; Brar, G.A.; Stern-Ginossar, N.; Harris, M.S.; Talhouarne, G.J.; Jackson, S.E.; Wills, M.R.; Weissman, J.S. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014, 8, 1365–1379. [Google Scholar] [CrossRef] [Green Version]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [Green Version]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezkurdia, I.; Juan, D.; Rodriguez, J.M.; Frankish, A.; Diekhans, M.; Harrow, J.; Vazquez, J.; Valencia, A.; Tress, M.L. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum. Mol. Genet. 2014, 23, 5866–5878. [Google Scholar] [CrossRef] [Green Version]
- Uchida, S.; Dimmeler, S. Long noncoding RNAs in cardiovascular diseases. Circ. Res. 2015, 116, 737–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thum, T.; Condorelli, G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ. Res. 2015, 116, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Schonrock, N.; Harvey, R.P.; Mattick, J.S. Long noncoding RNAs in cardiac development and pathophysiology. Circ. Res. 2012, 111, 1349–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viereck, J.; Thum, T. Circulating Noncoding RNAs as Biomarkers of Cardiovascular Disease and Injury. Circ. Res. 2017, 120, 381–399. [Google Scholar] [CrossRef]
- Ounzain, S.; Pedrazzini, T. The promise of enhancer-associated long noncoding RNAs in cardiac regeneration. Trends Cardiovasc. Med. 2015, 25, 592–602. [Google Scholar] [CrossRef] [Green Version]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [Green Version]
- Klattenhoff, C.A.; Scheuermann, J.C.; Surface, L.E.; Bradley, R.K.; Fields, P.A.; Steinhauser, M.L.; Ding, H.; Butty, V.L.; Torrey, L.; Haas, S.; et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013, 152, 570–583. [Google Scholar] [CrossRef] [Green Version]
- Ounzain, S.; Micheletti, R.; Arnan, C.; Plaisance, I.; Cecchi, D.; Schroen, B.; Reverter, F.; Alexanian, M.; Gonzales, C.; Ng, S.Y.; et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 2015, 89, 98–112. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Xu, Y.; Wang, Z.; Wu, Y.; Chen, J.; Wang, G.; Lu, C.; Jia, W.; Xi, J.; Zhu, S.; et al. A Linc1405/Eomes Complex Promotes Cardiac Mesoderm Specification and Cardiogenesis. Cell Stem Cell 2018, 22, 893–908. [Google Scholar] [CrossRef] [Green Version]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Wahrisch, S.; Beisaw, A.; Macura, K.; Blass, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gore-Panter, S.R.; Hsu, J.; Barnard, J.; Moravec, C.S.; Van Wagoner, D.R.; Chung, M.K.; Smith, J.D. PANCR, the PITX2 Adjacent Noncoding RNA, Is Expressed in Human Left Atria and Regulates PITX2c Expression. Circ. Arrhythm. Electrophysiol. 2016, 9, e003197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welsh, I.C.; Kwak, H.; Chen, F.L.; Werner, M.; Shopland, L.S.; Danko, C.G.; Lis, J.T.; Zhang, M.; Martin, J.F.; Kurpios, N.A. Chromatin Architecture of the Pitx2 Locus Requires CTCF- and Pitx2-Dependent Asymmetry that Mirrors Embryonic Gut Laterality. Cell Rep. 2015, 13, 337–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, K.M.; Anderson, D.M.; McAnally, J.R.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 2016, 539, 433–436. [Google Scholar] [CrossRef] [Green Version]
- Ritter, N.; Ali, T.; Kopitchinski, N.; Schuster, P.; Beisaw, A.; Hendrix, D.A.; Schulz, M.H.; Muller-McNicoll, M.; Dimmeler, S.; Grote, P. The lncRNA Locus Handsdown Regulates Cardiac Gene Programs and Is Essential for Early Mouse Development. Dev. Cell 2019, 50, 644–657. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Z.; Wu, C.; Pan, Z.; Xiang, L.; Liu, H.; Jin, X.; Tong, K.; Fan, S.; Jin, X. Potential association of long noncoding RNA HA117 with tetralogy of Fallot. Genes Dis. 2018, 5, 185–190. [Google Scholar] [CrossRef]
- Han, P.; Li, W.; Lin, C.H.; Yang, J.; Shang, C.; Nuernberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.Y.; Lin, C.J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Xu, Y.; Liang, C.; Xing, W.; Zhang, T. The mechanism of myocardial hypertrophy regulated by the interaction between mhrt and myocardin. Cell. Signal. 2018, 43, 11–20. [Google Scholar] [CrossRef]
- Viereck, J.; Kumarswamy, R.; Foinquinos, A.; Xiao, K.; Avramopoulos, P.; Kunz, M.; Dittrich, M.; Maetzig, T.; Zimmer, K.; Remke, J.; et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 2016, 8, 326ra22. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, X.J.; Ji, Y.X.; Zhang, P.; Deng, K.Q.; Gong, J.; Ren, S.; Wang, X.; Chen, I.; Wang, H.; et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016, 22, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Wo, Y.; Guo, J.; Li, P.; Yang, H.; Wo, J. Long non-coding RNA CHRF facilitates cardiac hypertrophy through regulating Akt3 via miR-93. Cardiovasc. Pathol. 2018, 35, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, F.; Zhou, L.Y.; Long, B.; Yuan, S.M.; Wang, Y.; Liu, C.Y.; Sun, T.; Zhang, X.J.; Li, P.F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014, 114, 1377–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.H.; Yuan, Y.X.; Rao, S.L.; Wang, P. LncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3653–3660. [Google Scholar]
- Li, Y.; Wang, J.; Sun, L.; Zhu, S. LncRNA myocardial infarction-associated transcript (MIAT) contributed to cardiac hypertrophy by regulating TLR4 via miR-93. Eur. J. Pharmacol. 2018, 818, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; An, X.; Li, Z.; Song, Y.; Li, L.; Zuo, S.; Liu, N.; Yang, G.; Wang, H.; Cheng, X.; et al. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 2016, 111, 56–65. [Google Scholar] [CrossRef]
- Jiang, F.; Zhou, X.; Huang, J. Long Non-Coding RNA-ROR Mediates the Reprogramming in Cardiac Hypertrophy. PLoS ONE 2016, 11, e0152767. [Google Scholar] [CrossRef] [Green Version]
- Lai, Y.; He, S.; Ma, L.; Lin, H.; Ren, B.; Ma, J.; Zhu, X.; Zhuang, S. HOTAIR functions as a competing endogenous RNA to regulate PTEN expression by inhibiting miR-19 in cardiac hypertrophy. Mol. Cell. Biochem. 2017, 432, 179–187. [Google Scholar] [CrossRef]
- Chen, H.; Cai, K. DSCAM-AS1 mediates pro-hypertrophy role of GRK2 in cardiac hypertrophy aggravation via absorbing miR-188-5p. Vitr. Cell. Dev. Biol. Anim. 2020, 56, 286–295. [Google Scholar] [CrossRef]
- Lv, L.; Li, T.; Li, X.; Xu, C.; Liu, Q.; Jiang, H.; Li, Y.; Liu, Y.; Yan, H.; Huang, Q.; et al. The lncRNA Plscr4 Controls Cardiac Hypertrophy by Regulating miR-214. Mol. Ther. Nucleic Acids 2018, 10, 387–397. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Liu, X.; Chen, L.; Chen, W.; Zhang, Y.; Chen, J.; Wu, X.; Zhao, Y.; Wu, X.; Sun, G. The long noncoding RNA XIST protects cardiomyocyte hypertrophy by targeting miR-330-3p. Biochem. Biophys. Res. Commun. 2018, 505, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Gu, Y.; Sun, Y.; Chen, J.; Wang, X.; Zhang, Y.; Gao, L.; Li, L. The long noncoding RNA XIST regulates cardiac hypertrophy by targeting miR-101. J. Cell. Physiol. 2019, 234, 13680–13692. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, R.; Yang, W.; Qi, B. SP1-SYNE1-AS1-miR-525-5p feedback loop regulates Ang-II-induced cardiac hypertrophy. J. Cell. Physiol. 2019, 234, 14319–14329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, F.; Wang, F.; Wu, N. Long noncoding RNA MAGI1-IT1 regulates cardiac hypertrophy by modulating miR-302e/DKK1/Wnt/beta-catenin signaling pathway. J. Cell. Physiol. 2020, 235, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Zhang, Y.H.; Li, R.B.; Zhou, L.Y.; An, T.; Zhang, R.C.; Zhai, M.; Huang, Y.; Yan, K.W.; Dong, Y.H.; et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat. Commun. 2018, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Zaccagnini, G.; Fuschi, P.; Voellenkle, C.; Carrara, M.; Sadeghi, I.; Bearzi, C.; Maimone, B.; Castelvecchio, S.; Stellos, K.; et al. Increased BACE1-AS long noncoding RNA and beta-amyloid levels in heart failure. Cardiovasc. Res. 2017, 113, 453–463. [Google Scholar] [CrossRef]
- Wang, K.; Long, B.; Zhou, L.Y.; Liu, F.; Zhou, Q.Y.; Liu, C.Y.; Fan, Y.Y.; Li, P.F. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 2014, 5, 3596. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Liu, C.Y.; Zhou, L.Y.; Wang, J.X.; Wang, M.; Zhao, B.; Zhao, W.K.; Xu, S.J.; Fan, L.H.; Zhang, X.J.; et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 2015, 6, 6779. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Liu, F.; Liu, C.Y.; An, T.; Zhang, J.; Zhou, L.Y.; Wang, M.; Dong, Y.H.; Li, N.; Gao, J.N.; et al. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ. 2016, 23, 1394–1405. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Zhao, Z.A.; Liu, J.; Hao, K.; Yu, Y.; Han, X.; Li, J.; Wang, Y.; Lei, W.; Dong, N.; et al. Long noncoding RNA Meg3 regulates cardiomyocyte apoptosis in myocardial infarction. Gene Ther. 2018, 25, 511–523. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, F.; Ma, W.; Zhang, P. Suppression of long noncoding RNA NEAT1 attenuates hypoxia-induced cardiomyocytes injury by targeting miR-378a-3p. Gene 2020, 731, 144324. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yang, X.S.; Zhang, Q.; Zhuang, X.; Dong, X.K.; Jiang, Y.H.; Tao, Y.N.; Yang, C.H. Downregulated LINC01614 Ameliorates Hypoxia/Reoxygenation-Stimulated Myocardial Injury by Directly Sponging microRNA-138-5p. Dose Response 2020, 18, 1559325820913786. [Google Scholar] [CrossRef] [Green Version]
- Lin, B.; Xu, J.; Wang, F.; Wang, J.; Zhao, H.; Feng, D. LncRNA XIST promotes myocardial infarction by regulating FOS through targeting miR-101a-3p. Aging 2020, 12, 7232–7247. [Google Scholar] [CrossRef] [PubMed]
- Micheletti, R.; Plaisance, I.; Abraham, B.J.; Sarre, A.; Ting, C.C.; Alexanian, M.; Maric, D.; Maison, D.; Nemir, M.; Young, R.A.; et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piccoli, M.T.; Gupta, S.K.; Viereck, J.; Foinquinos, A.; Samolovac, S.; Kramer, F.L.; Garg, A.; Remke, J.; Zimmer, K.; Batkai, S.; et al. Inhibition of the Cardiac Fibroblast-Enriched lncRNA Meg3 Prevents Cardiac Fibrosis and Diastolic Dysfunction. Circ. Res. 2017, 121, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhang, L.; Song, J.; Wang, Z.; Huang, X.; Guo, Z.; Chen, F.; Zhao, X. Long noncoding RNA MALAT1 mediates cardiac fibrosis in experimental postinfarct myocardium mice model. J. Cell. Physiol. 2019, 234, 2997–3006. [Google Scholar] [CrossRef]
- Qu, X.; Du, Y.; Shu, Y.; Gao, M.; Sun, F.; Luo, S.; Yang, T.; Zhan, L.; Yuan, Y.; Chu, W.; et al. MIAT Is a Pro-fibrotic Long Non-coding RNA Governing Cardiac Fibrosis in Post-infarct Myocardium. Sci. Rep. 2017, 7, 42657. [Google Scholar] [CrossRef]
- Wang, X.; Yong, C.; Yu, K.; Yu, R.; Zhang, R.; Yu, L.; Li, S.; Cai, S. Long Noncoding RNA (lncRNA) n379519 Promotes Cardiac Fibrosis in Post-Infarct Myocardium by Targeting miR-30. Med. Sci. Monit. 2018, 24, 3958–3965. [Google Scholar] [CrossRef]
- Zheng, D.; Zhang, Y.; Hu, Y.; Guan, J.; Xu, L.; Xiao, W.; Zhong, Q.; Ren, C.; Lu, J.; Liang, J.; et al. Long noncoding RNA Crnde attenuates cardiac fibrosis via Smad3-Crnde negative feedback in diabetic cardiomyopathy. FEBS J. 2019, 286, 1645–1655. [Google Scholar] [CrossRef] [Green Version]
- Zhu, P.; Yang, M.; Ren, H.; Shen, G.; Chen, J.; Zhang, J.; Liu, J.; Sun, C. Long noncoding RNA MALAT1 downregulates cardiac transient outward potassium current by regulating miR-200c/HMGB1 pathway. J. Cell. Biochem. 2018, 119, 10239–10249. [Google Scholar] [CrossRef]
- Long, Q.Q.; Wang, H.; Gao, W.; Fan, Y.; Li, Y.F.; Ma, Y.; Yang, Y.; Shi, H.J.; Chen, B.R.; Meng, H.Y.; et al. Long Noncoding RNA Kcna2 Antisense RNA Contributes to Ventricular Arrhythmias via Silencing Kcna2 in Rats With Congestive Heart Failure. J. Am. Heart Assoc. 2017, 6, e005965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Sun, L.; Xuan, L.; Pan, Z.; Hu, X.; Liu, H.; Bai, Y.; Jiao, L.; Li, Z.; Cui, L.; et al. Long non-coding RNA CCRR controls cardiac conduction via regulating intercellular coupling. Nat. Commun. 2018, 9, 4176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Jiao, L.; Sun, L.; Li, Y.; Gao, Y.; Xu, C.; Shao, Y.; Li, M.; Li, C.; Lu, Y.; et al. LncRNA ZFAS1 as a SERCA2a Inhibitor to Cause Intracellular Ca(2+) Overload and Contractile Dysfunction in a Mouse Model of Myocardial Infarction. Circ. Res. 2018, 122, 1354–1368. [Google Scholar] [CrossRef] [PubMed]
- Magny, E.G.; Pueyo, J.I.; Pearl, F.M.; Cespedes, M.A.; Niven, J.E.; Bishop, S.A.; Couso, J.P. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 2013, 341, 1116–1120. [Google Scholar] [CrossRef]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.J.; Lee, J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef] [Green Version]
- Buzin, C.H.; Mann, J.R.; Singer-Sam, J. Quantitative RT-PCR assays show Xist RNA levels are low in mouse female adult tissue, embryos and embryoid bodies. Development 1994, 120, 3529–3536. [Google Scholar]
- Gomez, J.A.; Wapinski, O.L.; Yang, Y.W.; Bureau, J.F.; Gopinath, S.; Monack, D.M.; Chang, H.Y.; Brahic, M.; Kirkegaard, K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 2013, 152, 743–754. [Google Scholar] [CrossRef] [Green Version]
- Kataoka, M.; Huang, Z.P.; Wang, D.Z. Build a braveheart: The missing linc (RNA). Circ. Res. 2013, 112, 1532–1534. [Google Scholar] [CrossRef] [Green Version]
- Franco, D.; Christoffels, V.M.; Campione, M. Homeobox transcription factor Pitx2: The rise of an asymmetry gene in cardiogenesis and arrhythmogenesis. Trends Cardiovasc. Med. 2014, 24, 23–31. [Google Scholar] [CrossRef]
- Essner, J.J.; Branford, W.W.; Zhang, J.; Yost, H.J. Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development 2000, 127, 1081–1093. [Google Scholar]
- Han, X.; Zhang, J.; Liu, Y.; Fan, X.; Ai, S.; Luo, Y.; Li, X.; Jin, H.; Luo, S.; Zheng, H.; et al. The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development 2019, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luther, H.P.; Haase, H.; Hohaus, A.; Beckmann, G.; Reich, J.; Morano, I. Characterization of naturally occurring myosin heavy chain antisense mRNA in rat heart. J. Cell. Biochem. 1998, 70, 110–120. [Google Scholar] [CrossRef]
- Yang, K.C.; Yamada, K.A.; Patel, A.Y.; Topkara, V.K.; George, I.; Cheema, F.H.; Ewald, G.A.; Mann, D.L.; Nerbonne, J.M. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 2014, 129, 1009–1021. [Google Scholar] [CrossRef]
- Li, H.; Chen, C.; Fan, J.; Yin, Z.; Ni, L.; Cianflone, K.; Wang, Y.; Wang, D.W. Identification of cardiac long non-coding RNA profile in human dilated cardiomyopathy. Cardiovasc. Res. 2018, 114, 747–758. [Google Scholar] [CrossRef]
- Saddic, L.A.; Sigurdsson, M.I.; Chang, T.W.; Mazaika, E.; Heydarpour, M.; Shernan, S.K.; Seidman, C.E.; Seidman, J.G.; Aranki, S.F.; Body, S.C.; et al. The Long Noncoding RNA Landscape of the Ischemic Human Left Ventricle. Circ. Cardiovasc. Genet. 2017, 10, 10. [Google Scholar] [CrossRef]
- Zangrando, J.; Zhang, L.; Vausort, M.; Maskali, F.; Marie, P.Y.; Wagner, D.R.; Devaux, Y. Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genom. 2014, 15, 460. [Google Scholar] [CrossRef] [Green Version]
- Ounzain, S.; Micheletti, R.; Beckmann, T.; Schroen, B.; Alexanian, M.; Pezzuto, I.; Crippa, S.; Nemir, M.; Sarre, A.; Johnson, R.; et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur. Heart J. 2015, 36, 353–368. [Google Scholar] [CrossRef]
- Nakamura, M.; Sadoshima, J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 2018, 15, 387–407. [Google Scholar] [CrossRef]
- Liu, J.; Wang, D.Z. An epigenetic “LINK(RNA)” to pathological cardiac hypertrophy. Cell Metab. 2014, 20, 555–557. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Liu, Y.; Guo, X.; Sun, G.; Ma, Q.; Dai, Y.; Zhu, G.; Sun, Y. Long noncoding RNA myocardial infarctionassociated transcript is associated with the microRNA1505p/P300 pathway in cardiac hypertrophy. Int. J. Mol. Med. 2018, 42, 1265–1272. [Google Scholar] [CrossRef] [Green Version]
- Prabhu, S.D.; Frangogiannis, N.G. The Biological Basis for Cardiac Repair After Myocardial Infarction: From Inflammation to Fibrosis. Circ. Res. 2016, 119, 91–112. [Google Scholar] [CrossRef]
- Jellis, C.; Martin, J.; Narula, J.; Marwick, T.H. Assessment of nonischemic myocardial fibrosis. J. Am. Coll. Cardiol. 2010, 56, 89–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murtha, L.A.; Schuliga, M.J.; Mabotuwana, N.S.; Hardy, S.A.; Waters, D.W.; Burgess, J.K.; Knight, D.A.; Boyle, A.J. The Processes and Mechanisms of Cardiac and Pulmonary Fibrosis. Front. Physiol. 2017, 8, 777. [Google Scholar] [CrossRef] [Green Version]
- Disertori, M.; Mase, M.; Ravelli, F. Myocardial fibrosis predicts ventricular tachyarrhythmias. Trends Cardiovasc. Med. 2017, 27, 363–372. [Google Scholar] [CrossRef]
- Wu, G.; Cai, J.; Han, Y.; Chen, J.; Huang, Z.P.; Chen, C.; Cai, Y.; Huang, H.; Yang, Y.; Liu, Y.; et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 2014, 130, 1452–1465. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.P.; Ding, Y.; Chen, J.; Wu, G.; Kataoka, M.; Hu, Y.; Yang, J.H.; Liu, J.; Drakos, S.G.; Selzman, C.H.; et al. Long non-coding RNAs link extracellular matrix gene expression to ischemic cardiomyopathy. Cardiovasc. Res. 2016, 112, 543–554. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Deng, J.; Xu, J.; Wang, H.; Yuan, M.; Liu, N.; Jiang, Y.; Liu, J. High-mobility group box 1 (HMGB1) downregulates cardiac transient outward potassium current (Ito) through downregulation of Kv4.2 and Kv4.3 channel transcripts and proteins. J. Mol. Cell. Cardiol. 2010, 49, 438–448. [Google Scholar] [CrossRef]
- Chilley, P.M.; Casson, S.A.; Tarkowski, P.; Hawkins, N.; Wang, K.L.; Hussey, P.J.; Beale, M.; Ecker, J.R.; Sandberg, G.K.; Lindsey, K. The POLARIS peptide of Arabidopsis regulates auxin transport and root growth via effects on ethylene signaling. Plant Cell 2006, 18, 3058–3072. [Google Scholar] [CrossRef] [Green Version]
- Bi, P.; Ramirez-Martinez, A.; Li, H.; Cannavino, J.; McAnally, J.R.; Shelton, J.M.; Sanchez-Ortiz, E.; Bassel-Duby, R.; Olson, E.N. Control of muscle formation by the fusogenic micropeptide myomixer. Science 2017, 356, 323–327. [Google Scholar] [CrossRef] [Green Version]
- Stein, C.S.; Jadiya, P.; Zhang, X.; McLendon, J.M.; Abouassaly, G.M.; Witmer, N.H.; Anderson, E.J.; Elrod, J.W.; Boudreau, R.L. Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep. 2018, 23, 3710–3720. [Google Scholar] [CrossRef]
- Nattel, S.; Harada, M. Atrial remodeling and atrial fibrillation: Recent advances and translational perspectives. J. Am. Coll. Cardiol. 2014, 63, 2335–2345. [Google Scholar] [CrossRef] [Green Version]
- Ke, Z.P.; Xu, Y.J.; Wang, Z.S.; Sun, J. RNA sequencing profiling reveals key mRNAs and long noncoding RNAs in atrial fibrillation. J. Cell. Biochem. 2019. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Guo, H.; Song, Y.; Chang, H.; Wang, S.; Zhang, M.; Liu, C. Long noncoding RNA AK055347 is upregulated in patients with atrial fibrillation and regulates mitochondrial energy production in myocardiocytes. Mol. Med. Rep. 2016, 14, 5311–5317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Ma, Z.; Guo, Z.; Zheng, M.; Li, K.; Yang, X. Analysis of long non-coding RNA and mRNA profiles in epicardial adipose tissue of patients with atrial fibrillation. Biomed. Pharmacother. 2020, 121, 109634. [Google Scholar] [CrossRef]
- Cao, F.; Li, Z.; Ding, W.M.; Yan, L.; Zhao, Q.Y. LncRNA PVT1 regulates atrial fibrosis via miR-128-3p-SP1-TGF-beta1-Smad axis in atrial fibrillation. Mol. Med. 2019, 25, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Xu, F.Q.; Guo, J.J.; Lin, P.L.; Meng, Z.; Hu, L.G.; Li, J.; Li, D.; Lu, X.H.; An, Y. Long noncoding RNA GAS5 attenuates cardiac fibroblast proliferation in atrial fibrillation via repressing ALK5. Eur. Rev. Med. Pharmacol Sci. 2019, 23, 7605–7610. [Google Scholar] [CrossRef]
- Chen, Q.; Feng, C.; Liu, Y.; Li, Q.F.; Qiu, F.Y.; Wang, M.H.; Shen, Z.D.; Fu, G.S. Long non-coding RNA PCAT-1 promotes cardiac fibroblast proliferation via upregulating TGF-beta1. Eur. Rev. Med. Pharmacol Sci. 2019, 23, 10517–10522. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Zhou, B.; You, L.; Hu, H.; Xie, R. LncRNA MIAT/miR-133a-3p axis regulates atrial fibrillation and atrial fibrillation-induced myocardial fibrosis. Mol. Biol. Rep. 2020, 47, 2605–2617. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Guo, Z.; Zhang, C.; Che, H.; Gong, W.; Shen, Z.; Shi, Y.; Ge, S. LncRNA NRON alleviates atrial fibrosis through suppression of M1 macrophages activated by atrial myocytes. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, X.; Zhang, Y.; Li, Z.; Xie, X.; Wang, J.; Gao, M.; Zhang, S.; Hou, Y. Transcriptome analysis of canine cardiac fat pads: Involvement of two novel long non-coding RNAs in atrial fibrillation neural remodeling. J. Cell. Biochem. 2015, 116, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.P.; Kirchhof, P. Pitx2 Adjacent Noncoding RNA: A New, Long, Noncoding Kid on the 4q25 Block. Circ. Arrhythm. Electrophysiol. 2016, 9, e003808. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, X.; Wang, W.; Du, J.; Wei, J.; Zhang, Y.; Wang, J.; Hou, Y. Altered long non-coding RNA expression profile in rabbit atria with atrial fibrillation: TCONS_00075467 modulates atrial electrical remodeling by sponging miR-328 to regulate CACNA1C. J. Mol. Cell. Cardiol. 2017, 108, 73–85. [Google Scholar] [CrossRef]
- Shen, C.; Kong, B.; Liu, Y.; Xiong, L.; Shuai, W.; Wang, G.; Quan, D.; Huang, H. YY1-induced upregulation of lncRNA KCNQ1OT1 regulates angiotensin II-induced atrial fibrillation by modulating miR-384b/CACNA1C axis. Biochem. Biophys. Res. Commun. 2018, 505, 134–140. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Lin, B.; Sheng, Y.; Yang, L. HBL1 Is a Human Long Noncoding RNA that Modulates Cardiomyocyte Development from Pluripotent Stem Cells by Counteracting MIR1. Dev. Cell 2017, 42, 333–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, Z.; Sun, X.; Sheng, H.; Zhu, L. Long non-coding RNA expression profile in atrial fibrillation. Int. J. Clin. Exp. Pathol. 2015, 8, 8402–8410. [Google Scholar]
- Mei, B.; Liu, H.; Yang, S.; Liang, M.Y.; Yue, Y.; Huang, S.Q.; Hou, J.; Chen, G.X.; Wu, Z.K. Long non-coding RNA expression profile in permanent atrial fibrillation patients with rheumatic heart disease. Eur. Rev. Med. Pharmacol Sci. 2018, 22, 6940–6947. [Google Scholar] [CrossRef]
- Wirka, R.C.; Gore, S.; Van Wagoner, D.R.; Arking, D.E.; Lubitz, S.A.; Lunetta, K.L.; Benjamin, E.J.; Alonso, A.; Ellinor, P.T.; Barnard, J.; et al. A common connexin-40 gene promoter variant affects connexin-40 expression in human atria and is associated with atrial fibrillation. Circ. Arrhythm. Electrophysiol. 2011, 4, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Gegonne, A.; Devaiah, B.N.; Singer, D.S. TAF7: Traffic controller in transcription initiation. Transcription 2013, 4, 29–33. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Han, D.; Shi, R.; Chen, M.; Sun, J.; Tian, H.; Yan, Y. Identification of atrial fibrillation-associated lncRNAs in atria from patients with rheumatic mitral valve disease. Microsc. Res. Tech. 2019, 82, 1136–1144. [Google Scholar] [CrossRef]
- Ling, L.E.; Lee, W.C. Tgf-beta type I receptor (Alk5) kinase inhibitors in oncology. Curr. Pharm. Biotechnol. 2011, 12, 2190–2202. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.J.; Zou, L.H.; Jin, J.H.; Xiao, F.; Li, L.; Liu, N.; Yang, J.F.; Zou, T. Long noncoding RNAs and novel inflammatory genes determined by RNA sequencing in human lymphocytes are up-regulated in permanent atrial fibrillation. Am. J. Transl. Res. 2017, 9, 2314–2326. [Google Scholar] [PubMed]
- Bettoni, M.; Zimmermann, M. Autonomic tone variations before the onset of paroxysmal atrial fibrillation. Circulation 2002, 105, 2753–2759. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Yuan, Y.; Qiu, C. Underexpression of CACNA1C Caused by Overexpression of microRNA-29a Underlies the Pathogenesis of Atrial Fibrillation. Med. Sci. Monit. 2016, 22, 2175–2181. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Zhang, Y.; Wang, N.; Pan, Z.; Gao, X.; Zhang, F.; Zhang, Y.; Shan, H.; Luo, X.; Bai, Y.; et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation 2010, 122, 2378–2387. [Google Scholar] [CrossRef]
- Nair, G.M.; Nery, P.B.; Redpath, C.J.; Birnie, D.H. The Role Of Renin Angiotensin System In Atrial Fibrillation. J. Atr. Fibrillation 2014, 6, 972. [Google Scholar] [CrossRef]
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grutzner, F.; Kaessmann, H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014, 505, 635–640. [Google Scholar] [CrossRef]
- Chodroff, R.A.; Goodstadt, L.; Sirey, T.M.; Oliver, P.L.; Davies, K.E.; Green, E.D.; Molnar, Z.; Ponting, C.P. Long noncoding RNA genes: Conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010, 11, R72. [Google Scholar] [CrossRef] [Green Version]
- Ng, S.Y.; Wong, C.K.; Tsang, S.Y. Differential gene expressions in atrial and ventricular myocytes: Insights into the road of applying embryonic stem cell-derived cardiomyocytes for future therapies. Am. J. Physiol. Cell Physiol. 2010, 299, C1234–C1249. [Google Scholar] [CrossRef] [Green Version]
- Del Alamo, J.C.; Lemons, D.; Serrano, R.; Savchenko, A.; Cerignoli, F.; Bodmer, R.; Mercola, M. High throughput physiological screening of iPSC-derived cardiomyocytes for drug development. Biochim. et Biophys. Acta (BBA) Mol. Cell Res. 2016, 1863, 1717–1727. [Google Scholar] [CrossRef]
- Devalla, H.D.; Schwach, V.; Ford, J.W.; Milnes, J.T.; El-Haou, S.; Jackson, C.; Gkatzis, K.; Elliott, D.A.; Chuva de Sousa Lopes, S.M.; Mummery, C.L.; et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015, 7, 394–410. [Google Scholar] [CrossRef] [PubMed]
- Ghazizadeh, Z.; Kiviniemi, T.; Olafsson, S.; Plotnick, D.; Beerens, M.E.; Zhang, K.; Gillon, L.; Steinbaugh, M.J.; Barrera, V.; Sui, S.H.; et al. Metastable Atrial State Underlies the Primary Genetic Substrate for MYL4 Mutation-Associated Atrial Fibrillation. Circulation 2020, 141, 301–312. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, J.; Han, P.; Yuan, Q.; Zhang, J.; Zhang, X.; Xu, Y.; Cao, H.; Meng, Q.; Chen, L.; et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011, 21, 579–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cyganek, L.; Tiburcy, M.; Sekeres, K.; Gerstenberg, K.; Bohnenberger, H.; Lenz, C.; Henze, S.; Stauske, M.; Salinas, G.; Zimmermann, W.H.; et al. Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes. JCI Insight 2018, 3, e99941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Protze, S.I.; Laksman, Z.; Backx, P.H.; Keller, G.M. Human Pluripotent Stem Cell-Derived Atrial and Ventricular Cardiomyocytes Develop from Distinct Mesoderm Populations. Cell Stem Cell 2017, 21, 179–194. [Google Scholar] [CrossRef]
- Argenziano, M.; Lambers, E.; Hong, L.; Sridhar, A.; Zhang, M.; Chalazan, B.; Menon, A.; Savio-Galimberti, E.; Wu, J.C.; Rehman, J.; et al. Electrophysiologic Characterization of Calcium Handling in Human Induced Pluripotent Stem Cell-Derived Atrial Cardiomyocytes. Stem Cell Rep. 2018, 10, 1867–1878. [Google Scholar] [CrossRef]
- Tanwar, V.; Bylund, J.B.; Hu, J.; Yan, J.; Walthall, J.M.; Mukherjee, A.; Heaton, W.H.; Wang, W.D.; Potet, F.; Rai, M.; et al. Gremlin 2 promotes differentiation of embryonic stem cells to atrial fate by activation of the JNK signaling pathway. Stem Cells 2014, 32, 1774–1788. [Google Scholar] [CrossRef] [Green Version]
- Bylund, J.B.; Trinh, L.T.; Awgulewitsch, C.P.; Paik, D.T.; Jetter, C.; Jha, R.; Zhang, J.; Nolan, K.; Xu, C.; Thompson, T.B.; et al. Coordinated Proliferation and Differentiation of Human-Induced Pluripotent Stem Cell-Derived Cardiac Progenitor Cells Depend on Bone Morphogenetic Protein Signaling Regulation by GREMLIN 2. Stem Cells Dev. 2017, 26, 678–693. [Google Scholar] [CrossRef] [Green Version]
- Laksman, Z.; Wauchop, M.; Lin, E.; Protze, S.; Lee, J.; Yang, W.; Izaddoustdar, F.; Shafaattalab, S.; Gepstein, L.; Tibbits, G.F.; et al. Modeling Atrial Fibrillation using Human Embryonic Stem Cell-Derived Atrial Tissue. Sci. Rep. 2017, 7, 5268. [Google Scholar] [CrossRef]
- Benzoni, P.; Campostrini, G.; Landi, S.; Bertini, V.; Marchina, E.; Iascone, M.; Ahlberg, G.; Olesen, M.S.; Crescini, E.; Mora, C.; et al. Human iPSC modelling of a familial form of atrial fibrillation reveals a gain of function of If and ICaL in patient-derived cardiomyocytes. Cardiovasc. Res. 2020, 116, 1147–1160. [Google Scholar] [CrossRef] [Green Version]
- HONG, L.; Zhang, M.; Youn, S.-W.; Lambers, E.; Sridhar, A.; Menon, A.; Chalazan, B.; Wu, J.C.; Rehman, J.; Darbar, D. Abstract 407: Modeling Atrial Fibrillation in a Dish Using Atrial iPSC Derived Cardiomyocytes. Circ. Res. 2019, 125, A407. [Google Scholar] [CrossRef]
- Girmatsion, Z.; Biliczki, P.; Bonauer, A.; Wimmer-Greinecker, G.; Scherer, M.; Moritz, A.; Bukowska, A.; Goette, A.; Nattel, S.; Hohnloser, S.H.; et al. Changes in microRNA-1 expression and IK1 up-regulation in human atrial fibrillation. Heart Rhythm. 2009, 6, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Zheng, S.; Xie, X.; Zhang, Y.; Wang, W.; Wang, Z.; Zhang, Y.; Wang, J.; Gao, M.; Hou, Y. MicroRNA-1 accelerates the shortening of atrial effective refractory period by regulating KCNE1 and KCNB2 expression: An atrial tachypacing rabbit model. PLoS ONE 2013, 8, e85639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celik, S.; Sadegh, M.K.; Morley, M.; Roselli, C.; Ellinor, P.T.; Cappola, T.; Smith, J.G.; Gidlof, O. Antisense regulation of atrial natriuretic peptide expression. JCI Insight 2019, 4, e130978. [Google Scholar] [CrossRef]
- Wood, E.J.; Chin-Inmanu, K.; Jia, H.; Lipovich, L. Sense-antisense gene pairs: Sequence, transcription, and structure are not conserved between human and mouse. Front. Genet. 2013, 4, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishimura, K.; Takeuchi, Y.; Fujiwara, K.; Tominaga, M.; Yoshioka, H.; Sawada, T. Quantitative analysis of the distribution of serotonin-immunoreactive cell bodies in the mouse brain. Neurosci. Lett. 1988, 91, 265–270. [Google Scholar] [CrossRef]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17, 556–565. [Google Scholar] [CrossRef] [Green Version]
- Marques, A.C.; Ponting, C.P. Catalogues of mammalian long noncoding RNAs: Modest conservation and incompleteness. Genome Biol. 2009, 10, R124. [Google Scholar] [CrossRef] [Green Version]
- Barichello, S.; Roberts, J.D.; Backx, P.; Boyle, P.M.; Laksman, Z. Personalizing therapy for atrial fibrillation: The role of stem cell and in silico disease models. Cardiovasc. Res. 2018, 114, 931–943. [Google Scholar] [CrossRef]
- Liu, J.; Laksman, Z.; Backx, P.H. The electrophysiological development of cardiomyocytes. Adv. Drug Deliv. Rev. 2016, 96, 253–273. [Google Scholar] [CrossRef]
- Blazeski, A.; Zhu, R.; Hunter, D.W.; Weinberg, S.H.; Boheler, K.R.; Zambidis, E.T.; Tung, L. Electrophysiological and contractile function of cardiomyocytes derived from human embryonic stem cells. Prog. Biophys. Mol. Biol. 2012, 110, 178–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danielsson, C.; Brask, J.; Skold, A.C.; Genead, R.; Andersson, A.; Andersson, U.; Stockling, K.; Pehrson, R.; Grinnemo, K.H.; Salari, S.; et al. Exploration of human, rat, and rabbit embryonic cardiomyocytes suggests K-channel block as a common teratogenic mechanism. Cardiovasc. Res. 2013, 97, 23–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, S.A.; Duff, J.; Bardi, I.; Zabielska, M.; Atanur, S.S.; Jabbour, R.J.; Simon, A.; Tomas, A.; Smolenski, R.T.; Harding, S.E.; et al. Biomimetic electromechanical stimulation to maintain adult myocardial slices in vitro. Nat. Commun. 2019, 10, 2168. [Google Scholar] [CrossRef] [PubMed]
- Ou, Q.; Jacobson, Z.; Abouleisa, R.R.E.; Tang, X.L.; Hindi, S.M.; Kumar, A.; Ivey, K.N.; Giridharan, G.; El-Baz, A.; Brittian, K.; et al. Physiological Biomimetic Culture System for Pig and Human Heart Slices. Circ. Res. 2019, 125, 628–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, C.; Milting, H.; Fein, E.; Reiser, E.; Lu, K.; Seidel, T.; Schinner, C.; Schwarzmayr, T.; Schramm, R.; Tomasi, R.; et al. Long-term functional and structural preservation of precision-cut human myocardium under continuous electromechanical stimulation in vitro. Nat. Commun. 2019, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Naito, H.; Melnychenko, I.; Didie, M.; Schneiderbanger, K.; Schubert, P.; Rosenkranz, S.; Eschenhagen, T.; Zimmermann, W.H. Optimizing engineered heart tissue for therapeutic applications as surrogate heart muscle. Circulation 2006, 114, 72–78. [Google Scholar] [CrossRef] [Green Version]
- Goldfracht, I.; Protze, S.; Shiti, A.; Setter, N.; Gruber, A.; Shaheen, N.; Nartiss, Y.; Keller, G.; Gepstein, L. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat. Commun. 2020, 11, 75. [Google Scholar] [CrossRef] [Green Version]

| Heart Development | ||
|---|---|---|
| lncRNA | Mechanism of Action | Study Model |
| Bvht [49] | Regulating cardiac mesoderm differentiation by targeting Mesp1 and SUZ12 | Mouse ESCs |
| CARMEN [50] | Contributing to cardiac specification by interacting with EZH2 and SUZ12 | Human CPCs; Mouse ESCs |
| lnc1405 [51] | Contributing to cardiogenesis by regulating Mesp1 transcription | Mouse ESCs and heart |
| Fendrr [52] | Regulating mesoderm specification by epigenetically silencing Foxf1 | Mouse ESCs |
| PANCR [53] | Positively regulating expression of PITX2, contributing left organ development | Human ESC-CMs |
| Playrr [54] | Negatively regulating expression of Pitx2, contributing right organ development | Mouse heart and ESCs; Chick heart |
| Uph [55] | Acting as upstream of Hand2 and positively regulates its expression | Mouse heart |
| Hdn [56] | Acting as downstream of Hand2 and negatively regulates its expression | Mouse heart |
| HA117 [57] | Suppressor of cardiac differentiation, linked with genetic disorder TOF | Patient |
| Cardiac Hypertrophy | ||
| Mhrt [58,59] | Protecting against hypertrophy by suppressing Brg1 and downregulating myocardin | Mouse heart (TAC); Rat CM |
| Chast [60] | Promoting hypertrophy by downregulating Plekhm1 | Mouse heart (TAC); hESC-CM (PE) |
| Chaer [61] | Promoting hypertrophy via suppressing inhibitory function of PRC2 on hypertrophy genes | Mouse heart (TAC) |
| CHRF [62,63] | Inducing hypertrophy by suppressing the anti-hypertrophic miR-489/Myd88 or miR-93/Akt3 axis | Mouse CM (AngII or Iso); Heart (AngII or TAC) |
| MIAT [64,65] | Inducing hypertrophy by suppressing the anti-hypertrophic miR-150/P300 or miR-93/TLR4 axis | Mouse heart or Rat H9c2 or Rat CM (AngII) |
| H19 [66] | Suppressing hypertrophy by targeting the miR-675/CaMKIIδ axis | Mouse heart (TAC); Mouse CM (PE) |
| lncRNA-ROR [67] | Promoting hypertrophy by targeting miR-133 | Mouse heart (TAC); Mouse CM (PE) |
| HOTAIR [68] | Suppressing hypertrophy by targeting miR-19 | Mouse heart (TAC); Mouse CM (AngII) |
| DSCAM-AS1 [69] | Boosting hypertrophy by targeting the miR-188-5p/GRK2 axis | Mouse CMs or Rat H9c2 (AngII) |
| Plscr4 [70] | Attenuating hypertrophy in vitro and in vivo by targeting the miR-214/Mfn2 axis | Mouse heart (TAC) or CM (AngII) |
| XIST [71,72] | Preventing hypertrophy in vitro and in vivo by targeting the miR-330-3p/S100B or miR-101/TLR2 axis | Mouse heart (TAC) or CM (PE); Rat H9c2 (PE) |
| SYNE1-AS1 [73] | Promoting hypertrophy in vitro and in vivo by targeting the miR-525-5p/SP1 axis | Mouse heart (TAC) or CM (AngII) |
| MAGI1-IT1 [74] | Suppressing hypertrophy by modulating the miR-302e/DKK1/Wnt/beta-catenin axis | Rat H9c2 (AngII) |
| Myocardial Infarction | ||
| CAIF [75] | Suppressing autophagy in infarcted CMs by blocking p53-mediated myocardin transcription | Mouse heart (I/R injury); CM (H2O2 injury) |
| BACE-AS1 [76] | Upregulating BACE1 transcripts that cause accumulation of β-amyloid and pathogenesis | Patient (ischemic HF); Mouse (MI) |
| CARL [77] | Suppressing mitochondrial fission and apoptosis through targeting the miR-359/PHB2 axis | Mouse heart (I/R injury) or CM (A/R) |
| APF [78] | Inducing adaptive cell autophagy through targeting the miR-188-3p/ATG7 axis | Mouse heart (I/R injury) or CM (A/R) |
| NRF [79] | Inducing cardiac necrosis through targeting the miR-873/RIPK1/RIPK3 axis | Mouse heart (I/R) or CM (H2O2) |
| Meg3 [80] | Inducing cardiomyocyte apoptosis by direct binding with RNA-binding protein FUS | Mouse heart (MI) or CM (hypoxia); hESC-CM (hypoxia) or HF Patient |
| NEAT1 [81] | Regulating CM proliferation through suppression of miR-378-3p | Rat Heart (I/R injury) or CM (hypoxia or H2O2); Patients (MI) |
| LINC01614 [82] | Promoting MI by suppression of miR-138-5p | Patient (MI); Rat H9c2 (H/R) |
| XIST [83] | Promoting apoptosis and MI by targeting miR-101-3p/FOS axis | Mouse Heart (MI) or CM (hypoxia) |
| Cardiac Fibrosis | ||
| WISPER [84] | Promoting fibroblast proliferation and differentiation through activation of TIA1-related protein | Mouse heart (MI) or CF; Human CF |
| Meg3 [85] | Promoting cardiac fibrosis through activation of the p53/MMP2 axis | Mouse heart (TAC) or CF (TGF-β1) |
| MALAT1 (NEAT2) [86] | Promoting fibroblast proliferation through targeting miR-145/ TGF-β1 axis | Mouse heart (MI) or CF (Ang-II) |
| MIAT [87] | Promoting cardiac fibrosis through targeting miR-24/Furin/ TGF-β1 axis | Mouse heart (MI) or CF (Ang-II) |
| n379519 [88] | Promoting cardiac fibrosis through targeting miR-30 | Rat heart (MI) or CF (TGF-β1) |
| Crnde [89] | Attenuating fibrosis via Smad3-Crnde negative feedback | Mouse heart (DCM) or CF (TGF-β1) |
| Cardiac Arrhythmias | ||
| MALAT1 [90] | Overexpression dysregulates Ito through targeting the miR-200c/HMGB1 axis | Rat heart (MI) or CM |
| Kcna-AS [91] | Contributing to ventricular arrhythmias by downregulating Kcna expression | Rat heart (TAC) or CM (PE); Patient (HF) |
| CCRR [92] | Increasing Cx43 levels to improve intercellular cardiac conduction | Mouse heart (TAC) or CM; Patient (HF) or AC16 |
| ZFAS1 [93] | Repressing SERCA2a expression to dysregulate Ca2+ homeostasis | Mouse heart (MI) or CM (hypoxia); Patient (MI) or AC16 (hypoxia) |
| pncr003:2 L [94] | Encoding micropeptide Sarcolamban, which regulates SERCA function | Drosophila heart |
| LOC100507537 [95] | Encoding micropeptide DWORF, which positively regulates SERCA activity | Mouse heart (MI) or CM; Patient (MI) |
| Development of AF | |||
|---|---|---|---|
| LncRNA | Expression in AF | Mechanism of Action | Study Model |
| RP11-99E15.2 [123] | ND | May be involved in AF by regulating extracellular matrix binding via interactions with ITGB3 | Patient (AF) |
| RP3-523K23.2 [123] | ND | May be involved in AF by regulating transcription of HSF2 | Patient (AF) |
| AK055347 [124] | Up | Dysregulating mitochondrial energy production by regulating mitochondrial Cyp450, ATP synthase, and MSS51 | Patient (AF) |
| Structural Remodeling | |||
| PVT1 [125,126] | Up | Regulating miR-128-3p/Sp1/TGF-β1/Smad axis by sponging miR-128-3p | Patient (AF) or Atrial fibroblast; Mouse heart (Ang-II) |
| GAS5 [127] | Down | Inhibiting ALK5 and suppresses cell proliferation | Patient (AF) or AC16 |
| PCAT1 [128] | Up | Promoting fibroblast proliferation through targeting TGF-β1 | Patient (AF) or AC16 |
| MIAT [129] | Up | Alleviating AF and reducing atrial fibrosis by suppressing miR-133-3p | Patient (AF); Rat (electrical stimulation) |
| NRON [130] | Up | Inhibiting NFAT localization to nucleus, thus suppresses IL-12 and macrophage switch from M2 to M1. | Mouse atrial CM (AngII) |
| TCONS_00032546 [131] | Down | Related to RAS-mediated neuronal remodeling in cardiac fat pads | Canine heart (atrial tachypacing) |
| TCONS_00026102 [131] | Down | Related to RAS-mediated neuronal remodeling in cardiac fat pads | Canine heart (atrial tachypacing) |
| Electrical Remodeling | |||
| PANCR [53,132] | ND | Regulating PITX2, an AF-related gene, but not studied in AF directly | Human ESC-CM |
| TCONS_00075467 [133] | Down | Upregulating of it results with increased sponging of miR328, thus increasing CACNA1C levels | Rabbit right atria (AF) |
| KCNQ1OT1 [134] | Up | Downregulating of it results with decreased sponging of miR384, thus decreasing CACNA1C levels | Mouse heart (AngII) or CM |
| NPPA-AS1 [123] | Up | Modulating cardiac contraction genes (e.g., NPPA, PLCE1, TNNC1, TNN1). | Patient (AF) |
| lncRNA-HBL1 [135] | Up | Downregulating miR-1, an AF-related gene, but not studied in AF directly | Human iPSC-CM |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bektik, E.; Cowan, D.B.; Wang, D.-Z. Long Non-Coding RNAs in Atrial Fibrillation: Pluripotent Stem Cell-Derived Cardiomyocytes as a Model System. Int. J. Mol. Sci. 2020, 21, 5424. https://doi.org/10.3390/ijms21155424
Bektik E, Cowan DB, Wang D-Z. Long Non-Coding RNAs in Atrial Fibrillation: Pluripotent Stem Cell-Derived Cardiomyocytes as a Model System. International Journal of Molecular Sciences. 2020; 21(15):5424. https://doi.org/10.3390/ijms21155424
Chicago/Turabian StyleBektik, Emre, Douglas B. Cowan, and Da-Zhi Wang. 2020. "Long Non-Coding RNAs in Atrial Fibrillation: Pluripotent Stem Cell-Derived Cardiomyocytes as a Model System" International Journal of Molecular Sciences 21, no. 15: 5424. https://doi.org/10.3390/ijms21155424
APA StyleBektik, E., Cowan, D. B., & Wang, D. -Z. (2020). Long Non-Coding RNAs in Atrial Fibrillation: Pluripotent Stem Cell-Derived Cardiomyocytes as a Model System. International Journal of Molecular Sciences, 21(15), 5424. https://doi.org/10.3390/ijms21155424