Dual Substrate Specificity of the Rutinosidase from Aspergillus niger and the Role of Its Substrate Tunnel
Abstract
:1. Introduction
2. Results
2.1. Production of Recombinant Rutinosidase from A. niger
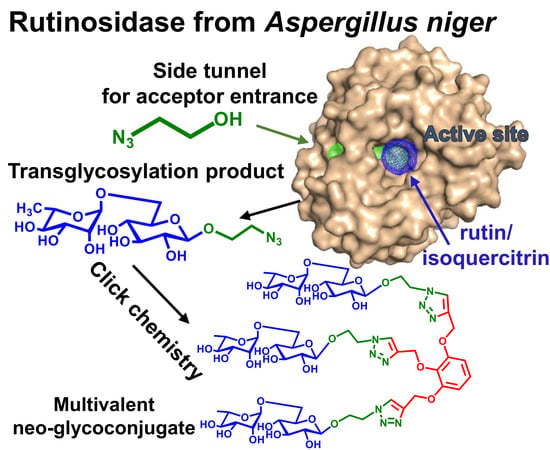
2.2. Transglycosylation Products
2.3. Product Stability against Enzymatic Hydrolysis
2.4. Application of Transglycosylation Products
2.5. AnRut Structure and Description of Substrate Tunnels
2.6. Dual Substrate Specificity and Substrate Docking
2.7. Acceptor Specificity of AnRut
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Recombinant Rutinosidase from A. niger
4.3. Enzyme Activity Assay
4.4. Analytical Transglycosylation Reactions
4.5. Preparative Transglycosylation Reactions
4.6. Product Stability against Enzymatic Hydrolysis
4.7. Structural Characterization
4.8. Synthesis of 1,2,3-tris-[1-(α-l-rhamnopyranosyl-(1→6)-β-d-glucopyranosyl)-1H-1,2,3-triazol-4-yl)-2-ethyloxy]benzene (25)
4.9. Modeling of Enzyme Tunnels and Substrate Docking
4.10. Molecular Dynamics Simulations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AnRut | Recombinant rutinosidase from Aspergillus niger |
| GH | Glycoside hydrolase |
| AGE | Advanced glycation end-products |
| SDS | Sodium dodecyl sulfate |
| TLC | Thin-layer chromatography |
| DMSO | Dimethyl sulfoxide |
| MS | Mass spectrometry |
| ESI-MS | Electrospray ionization mass spectrometry |
| HPLC | High-performance liquid chromatography |
| NMR | Nuclear magnetic resonance |
| PDB | Protein data bank |
| HRMS | High-resolution mass spectrometry |
| NVT | Constant number of particles, volume, and temperature |
| NPT | Constant number of particles, pressure, and temperature |
| RMSD | Root mean square deviation |
| RMSF | Root mean square fluctuations |
References
- Minig, M.; Mazzaferro, L.S.; Erra-Balsells, R.; Petroselli, G.; Breccia, J.D. α-Rhamnosyl-β-glucosidase-catalyzed reactions for analysis and biotransformations of plant-based foods. J. Agric. Food Chem. 2011, 59, 11238–11243. [Google Scholar] [CrossRef]
- Gunata, Z.; Blondeel, C.; Vallier, M.J.; Lepoutre, J.P.; Sapis, J.C.; Watanabe, N. An endoglycosidase from grape berry skin of cv. M. Alexandria hydrolyzing potentially aromatic disaccharide glycosides. J. Agric. Food Chem. 1998, 46, 2748–2753. [Google Scholar] [CrossRef]
- Neher, B.D.; Mazzaferro, L.S.; Kotik, M.; Oyhenart, J.; Halada, P.; Křen, V.; Breccia, J.D. Bacteria as source of diglycosidase activity: Actinoplanes missouriensis produces 6-O-α-l-rhamnosyl-β-d-glucosidase active on flavonoids. Appl. Microbiol. Biotechnol. 2016, 100, 3061–3070. [Google Scholar] [CrossRef] [PubMed]
- Weiz, G.; Mazzaferro, L.S.; Kotik, M.; Neher, B.D.; Halada, P.; Křen, V.; Breccia, J.D. The flavonoid degrading fungus Acremonium sp. DSM 24697 produces two diglycosidases with different specificities. Appl. Microbiol. Biotechnol. 2019, 103, 9493–9504. [Google Scholar] [CrossRef] [PubMed]
- Koseki, T.; Ishikawa, M.; Kawasaki, M.; Shiono, Y. β-Diglycosidases from microorganisms as industrial biocatalysts: Biochemical characteristics and potential applications. Appl. Microbiol. Biotechnol. 2018, 102, 8717–8723. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Schluesener, H. Health-promoting effects of the citrus flavanone hesperidin. Crit. Rev. Food Sci. Nutr. 2017, 57, 613–631. [Google Scholar] [CrossRef]
- Weng, S.S.; Mao, L.; Gong, Y.Y.; Sun, T.; Gu, Q. Role of quercetin in protecting ARPE-19 cells against H2O2-induced injury via nuclear factor erythroid 2 like 2 pathway activation and endoplasmic reticulum stress inhibition. Mol. Med. Rep. 2017, 16, 3461–3468. [Google Scholar] [CrossRef] [Green Version]
- Boadi, W.Y.; Lo, A. Effects of quercetin, kaempferol and exogenous glutathione on phospho- and total-AKT in 3T3-L1 preadipocytes. J. Diet. Suppl. 2018, 15, 814–826. [Google Scholar] [CrossRef]
- Rezvan, N.; Moini, A.; Gorgani-Firuzjaee, S.; Hosseinzadeh-Attar, M.J. Oral quercetin supplementation enhances adiponectin receptor transcript expression in polycystic ovary syndrome patients: A randomized placebo-controlled double-blind clinical trial. J. Diet. Suppl. 2018, 19, 627–633. [Google Scholar] [CrossRef]
- Nam, H.K.; Hong, S.H.; Shin, K.C.; Oh, D.K. Quercetin production from rutin by a thermostable β-rutinosidase from Pyrococcus furiosus. Biotechnol. Lett. 2012, 34, 483–489. [Google Scholar] [CrossRef]
- Mazzaferro, L.S.; Breccia, J.D. Functional and biotechnological insights into diglycosidases. J. Diet. Suppl. 2011, 29, 103–112. [Google Scholar] [CrossRef]
- Pageon, H.; Azouaoui, A.; Zucchi, H.; Ricois, S.; Tran, C.; Asselineau, D. Potentially beneficial effects of rhamnose on skin ageing: An in vitro and in vivo study. Int. J. Cosmet. Sci. 2019, 41, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Šimčíková, D.; Kotik, M.; Weignerová, L.; Halada, P.; Pelantová, H.; Adamcová, K.; Křen, V. α-l-Rhamnosyl-β-d-glucosidase (rutinosidase) from Aspergillus niger: Characterization and synthetic potential of a novel diglycosidase. Adv. Synth. Catal. 2015, 357, 107–117. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzyme database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [Green Version]
- Bassanini, I.; Kapešová, J.; Petrásková, L.; Pelantová, H.; Markošová, K.; Rebroš, M.; Valentová, K.; Kotik, M.; Káňová, K.; Bojarová, P.; et al. Glycosidase-catalyzed synthesis of glycosyl esters and phenolic glycosides of aromatic acids. Adv. Synth. Catal. 2019, 361, 2627–2637. [Google Scholar] [CrossRef] [Green Version]
- Bojarová, P.; Bruthans, J.; Křen, V. β-N-Acetylhexosaminidases—The wizards of glycosylation. Appl. Microbiol. Biotechnol. 2019, 103, 7869–7881. [Google Scholar] [CrossRef]
- Kapešová, J.; Petrásková, L.; Markošová, K.; Rebroš, M.; Kotik, M.; Bojarová, P.; Křen, V. Bioproduction of quercetin and rutinose catalyzed by rutinosidase: Novel concept of “solid state biocatalysis”. Int. J. Mol. Sci. 2019, 20, 1112. [Google Scholar] [CrossRef] [Green Version]
- Pachl, P.; Kapešová, J.; Brynda, J.; Biedermannová, L.; Pelantová, H.; Bojarová, P.; Křen, V.; Řezáčová, P.; Kotik, M. Rutinosidase from Aspergillus niger: Crystal structure and insight into the enzymatic activity. FEBS J. 2020, 287, 3315–3327. [Google Scholar] [CrossRef]
- Katayama, S.; Ohno, F.; Yamauchi, Y.; Kato, M.; Makabe, H.; Nakamura, S. Enzymatic synthesis of novel phenol acid rutinosides using rutinase and their antiviral activity in vitro. J. Agric. Food Chem. 2013, 61, 9617–9622. [Google Scholar] [CrossRef]
- Houlmont, J.P.; Vercruysse, K.; Perez, E.; Rico-Lattes, I.; Bordat, P.; Treilhou, M. Cosmetic use formulations containing pentyl rhamnoside and cetyl rhamnoside. Int. J. Cosmet. Sci. 2001, 23, 363–368. [Google Scholar] [CrossRef]
- Traber, P.G.; Chou, H.; Zomer, E.; Hong, F.; Klyosov, A.; Fiel, M.I.; Friedman, S.L. Regression of fibrosis and reversal of cirrhosis in rats by galectin inhibitors in thioacetamide-induced liver disease. PLoS ONE 2013, 8, e75361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bu, L.T.; Beckham, G.T.; Shirts, M.R.; Nimlos, M.R.; Adney, W.S.; Himmel, M.E.; Crowley, M.F. Probing carbohydrate product expulsion from a processive cellulase with multiple absolute binding free energy methods. J. Biol. Chem. 2011, 286, 18161–18169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varrot, A.; Frandsen, T.P.; von Ossowski, I.; Boyer, V.; Cottaz, S.; Driguez, H.; Schulein, M.; Davies, G.J. Structural basis for ligand binding and processivity in cellobiohydrolase Cel6A from Humicola insolens. Structure 2003, 11, 855–864. [Google Scholar] [CrossRef] [Green Version]
- Cutfield, S.M.; Davies, G.J.; Murshudov, G.; Anderson, B.F.; Moody, P.C.E.; Sullivan, P.A.; Cutfield, J.F. The structure of the exo-β-(1,3)-glucanase from Candida albicans in native and bound forms: Relationship between a pocket and groove in family 5 glycosyl hydrolases. J. Mol. Biol. 1999, 294, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Ferguson, A.D.; Bergeron, J.J.M.; Thomas, D.Y. The ER protein folding sensor UDP-glucose glycoprotein-glucosyltransferase modifies substrates distant to local changes in glycoprotein conformation. Nat. Struct. Mol. Biol. 2004, 11, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Kotik, M.; Brodsky, K.; Halada, P.; Javůrková, H.; Pelantová, H.; Konvalinková, D.; Bojarová, P.; Křen, V. Access to both anomers of rutinosyl azide using wild-type rutinosidase and its catalytic nucleophile mutant. Unpublished work. 2020. [Google Scholar]
- Ly, H.D.; Withers, S.G. Mutagenesis of glycosidases. Annu. Rev. Biochem. 1999, 68, 487–522. [Google Scholar] [CrossRef]
- Bojarová, P.; Kulik, N.; Slámová, K.; Hubálek, M.; Kotik, M.; Cvačka, J.; Pelantová, H.; Křen, V. Selective β-N-acetylhexosaminidase from Aspergillus versicolor—A tool for producing bioactive carbohydrates. Appl. Microbiol. Biotechnol. 2019, 103, 1737–1753. [Google Scholar] [CrossRef]
- Chovancová, E.; Pavelka, A.; Beneš, P.; Strnad, O.; Březovský, J.; Kozliková, B.; Gora, A.; Sustr, V.; Klvaňa, M.; Medek, P.; et al. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 2012, 8, e1002708. [Google Scholar] [CrossRef] [Green Version]
- Gouaux, E.; MacKinnon, R. Principles of selective ion transport in channels and pumps. Science 2005, 310, 1461–1465. [Google Scholar] [CrossRef] [Green Version]
- Klvaňa, M.; Pavlová, M.; Koudeláková, T.; Chaloupková, R.; Dvořák, P.; Prokop, Z.; Stsiapanava, A.; Kutý, M.; Kutá-Smatanová, I.; Dohnálek, J.; et al. Pathways and mechanisms for product release in the engineered haloalkane dehalogenases explored using classical and random acceleration molecular dynamics simulations. J. Mol. Biol. 2009, 392, 1339–1356. [Google Scholar] [CrossRef]
- Zámocký, M.; Herzog, C.; Nykyri, L.M.; Koller, F. Site-directed mutagenesis of the lower parts of the major substrate channel of yeast catalase A leads to highly increased peroxidatic activity. FEBS Lett. 1995, 367, 241–245. [Google Scholar] [CrossRef] [Green Version]
- Wen, Z.; Baudry, J.; Berenbaum, M.R.; Schuler, M.A. Ile115Leu mutation in the SRS1 region of an insect cytochrome P450 (CYP6B1) compromises substrate turnover via changes in a predicted product release channel. Protein Eng. Des. Sel. 2005, 18, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Stourac, J.; Vavra, O.; Kokkonen, P.; Filipovic, J.; Pinto, G.; Brezovsky, J.; Damborsky, J.; Bednar, D. Caver Web 1.0: Identification of tunnels and channels in proteins and analysis of ligand transport. Nucleic Acids Res. 2019, 47, W414–W422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patrick, W.M.; Nakatani, Y.; Cutfield, S.M.; Sharpe, M.L.; Ramsay, R.J.; Cutfield, J.F. Carbohydrate binding sites in Candida albicans exo-β-1,3-glucanase and the role of the Phe-Phe ‘clamp’ at the active site entrance. FEBS J. 2010, 277, 4549–4561. [Google Scholar] [CrossRef] [PubMed]
- Mazzaferro, L.S.; Pinuel, L.; Erra-Balsells, R.; Giudicessi, S.L.; Breccia, J.D. Transglycosylation specificity of Acremonium sp. α-rhamnosyl-β-glucosidase and its application to the synthesis of the new fluorogenic substrate 4-methylumbelliferyl-rutinoside. Carbohydr. Res. 2012, 347, 69–75. [Google Scholar] [CrossRef]
- Gerstorferová, D.; Fliedrová, B.; Halada, P.; Marhol, P.; Křen, V.; Weignerová, L. Recombinant α-l-rhamnosidase from Aspergillus terreus in selective trimming of rutin. Process Biochem. 2012, 47, 828–835. [Google Scholar] [CrossRef]
- Pavelka, A. Bioinformatics Tools for the Analysis of Macromolecular Structures. Ph.D Thesis, Masaryk University, Brno, Czech Republic, 2016. [Google Scholar]
- Brezovsky, J.; Babkova, P.; Degtjarik, O.; Fortova, A.; Gora, A.; Iermak, I.; Rezacova, P.; Dvorak, P.; Smatanova, I.K.; Prokop, Z.; et al. Engineering a de novo transport tunnel. ACS Catal. 2016, 6, 7597–7610. [Google Scholar] [CrossRef]
- Vavra, O.; Filipovic, J.; Plhak, J.; Bednar, D.; Marques, S.M.; Brezovsky, J.; Stourac, J.; Matyska, L.; Damborsky, J. CaverDock: A molecular docking-based tool to analyse ligand transport through protein tunnels and channels. Bioinformatics 2019, 35, 4986–4993. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Chen, J.; Cheng, T.J.; Gindulyte, A.; He, J.; He, S.Q.; Li, Q.L.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dassault Systèmes BIOVIA. Discovery Studio Modeling Environment, Release 2017; Dassault Systèmes: San Diego, CA, USA, 2016. [Google Scholar]
- Berendsen, H.J.C.; Vanderspoel, D.; Vandrunen, R. GROMACS—A message-passing parallel molecular-dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Lindahl, E.; Hess, B.; van der Spoel, D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Model. 2001, 7, 306–317. [Google Scholar] [CrossRef]







| Glycosyl Donor Acceptor | Rutin | Isoquercitrin | ||
|---|---|---|---|---|
| Product | Yield (%) | Product | Yield (%) | |
| pentanol (3) | 10 | 16 | 17 | 15 |
| sodium azide (4) | 11 | 44 | 18 | 22 |
| 2-azidoethanol (5) | 12 | 16 | 19 | 11 |
| 2-phenylethanol (6) | 13 | 9 | 20 | 7 |
| catechol (7) | 14b | <7 | 21 | 9 |
| p-nitrophenol (8) | 15 | 7 | 22 | 8 |
| 4-methylumbelliferone (9) | 16c | <5 | 23c | <5 |
| Rutinosides | Hydrolysis Rate | β-Glucosides | Hydrolysis Rate |
|---|---|---|---|
| 10 | - | 17 | - |
| 11 | - | 18 | - |
| 12 | - | 19 | - |
| 13 | - | 20 | - |
| 14 | + | 21 | ++ |
| 15 | ++ | 22 | ++ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brodsky, K.; Kutý, M.; Pelantová, H.; Cvačka, J.; Rebroš, M.; Kotik, M.; Kutá Smatanová, I.; Křen, V.; Bojarová, P. Dual Substrate Specificity of the Rutinosidase from Aspergillus niger and the Role of Its Substrate Tunnel. Int. J. Mol. Sci. 2020, 21, 5671. https://doi.org/10.3390/ijms21165671
Brodsky K, Kutý M, Pelantová H, Cvačka J, Rebroš M, Kotik M, Kutá Smatanová I, Křen V, Bojarová P. Dual Substrate Specificity of the Rutinosidase from Aspergillus niger and the Role of Its Substrate Tunnel. International Journal of Molecular Sciences. 2020; 21(16):5671. https://doi.org/10.3390/ijms21165671
Chicago/Turabian StyleBrodsky, Katerina, Michal Kutý, Helena Pelantová, Josef Cvačka, Martin Rebroš, Michael Kotik, Ivana Kutá Smatanová, Vladimír Křen, and Pavla Bojarová. 2020. "Dual Substrate Specificity of the Rutinosidase from Aspergillus niger and the Role of Its Substrate Tunnel" International Journal of Molecular Sciences 21, no. 16: 5671. https://doi.org/10.3390/ijms21165671
APA StyleBrodsky, K., Kutý, M., Pelantová, H., Cvačka, J., Rebroš, M., Kotik, M., Kutá Smatanová, I., Křen, V., & Bojarová, P. (2020). Dual Substrate Specificity of the Rutinosidase from Aspergillus niger and the Role of Its Substrate Tunnel. International Journal of Molecular Sciences, 21(16), 5671. https://doi.org/10.3390/ijms21165671