Tracking Antimicrobial Resistance Determinants in Diarrheal Pathogens: A Cross-Institutional Pilot Study
Abstract
:1. Introduction
2. Results and Discussion
2.1. Overall Population Characteristics
2.2. AMR Genotypes Detected in Each Genus
2.2.1. Campylobacter spp.
2.2.2. E. coli
2.2.3. Shigella spp.
2.2.4. Salmonella spp.
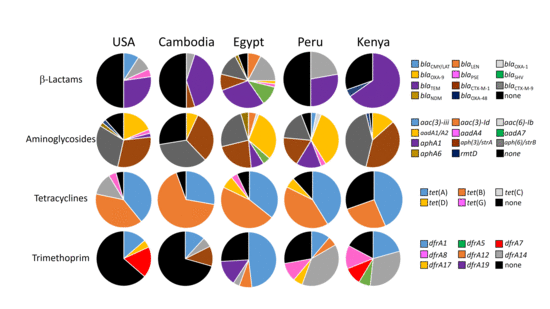
2.3. Geographic Trends
3. Materials and Methods
3.1. Isolates
3.2. Processing and DNA Hybridization
3.3. Confirmatory PCR
3.4. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AG | Aminoglycoside |
| AMP | Antimicrobial peptide |
| AMR | Antimicrobial resistance |
| ANS | Ansamycins |
| ARD | Antimicrobial resistance determinant |
| ARDM | Antimicrobial Resistance Determinant Microarray |
| AZM | Azithromycin |
| BLA | β-Lactamase |
| CDC | Centers for Disease Control and Prevention |
| CLSI | Clinical and Laboratory Standards Institute |
| ESBL | Extended-spectrum β-lactamase |
| FQ | Fluoroquinolone |
| GEIS | Global Emerging Infections Surveillance and Response System |
| GLY | Glycopeptides |
| LIN | Lincosamides |
| MAC | Macrolides |
| MLS | Macrolide/lincosamide/streptogramin |
| MUP | Mupirocin |
| NAMRU | Naval Medical Research Unit |
| NRL | Naval Research Laboratory |
| NTS | Non-typhoidal Salmonella |
| PHE | Phenicols |
| PMQR | Plasmid-mediated quinolone resistance |
| PT | Platensimycin + platencin |
| QUA | Quaternary amines |
| SGI | Salmonella genomic island |
| SNP | Single nucleotide polymorphism |
| SRL | Shigella resistance island |
| STR | Streptothricin |
| SUL | Sulfonamides |
| SXT | Trimethoprim-sulfamethoxazole |
| TET | Tetracycline |
| TMP | Diaminopyrimidine/trimethoprim |
| USAMRD | US Army Medical Research Directorate, Africa/Kenya Microbiology Hub, Kericho |
References
- Liu, L.; Johnson, H.L.; Cousens, S.; Perin, J.; Scott, S.; Lawn, J.E.; Rudan, I.; Campbell, H.; Cibulskis, R.; Li, M.; et al. Global, regional, and national causes of child mortality: An updated systematic analysis for 2010 with time trends since 2000. Lancet 2012, 379, 2151–2161. [Google Scholar] [CrossRef]
- James, S.L.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef] [Green Version]
- Reiner, R.C.; Blacker, B.F.; Khalil, I.A.; Rao, P.C.; Cao, S.; Zimsen, S.R.; Albertson, S.B.; Stanaway, J.D.; Deshpande, A.; Abebe, Z.; et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1211–1228. [Google Scholar] [CrossRef] [Green Version]
- Prüss-Ustün, A.; Bartram, J.; Clasen, T.; Colford, J.M., Jr.; Cumming, O.; Curtis, V.; Bonjour, S.; Dangour, A.D.; De France, J.; Fewtrell, L.; et al. Burden of disease from inadequate water, sanitation and hygiene in low- and middle-income settings: A retrospective analysis of data from 145 countries. Trop. Med. Int. Health 2014, 19, 894–905. [Google Scholar] [CrossRef] [Green Version]
- Cairncross, S.; Hunt, C.; Boisson, S.; Bostoen, K.; Curtis, V.; Fung, I.C.H.; Schmidt, W.P. Water, sanitation and hygiene for the prevention of diarrhoea. Int. J. Epidemiol. 2010, 39, i193–i205. [Google Scholar] [CrossRef] [Green Version]
- Araya, P.; Hug, J.; Joy, G.; Oschmann, F.; Rubinstein, S. The Impact of Water and Sanitation on Diarrhoeal Disease Burden and Over-Consumption of Antibiotics; London School of Economics and Political Science: London, UK, 2016. [Google Scholar]
- Casburn-Jones, A.C.; Farthing, M.J. Management of infectious diarrhea. Gut 2004, 53, 296–305. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. The Treatment for Diarrhoea: A Manual for Physicians and Other Senior Heath Workers; World Health Organization: Geneva, Switzerland, 2005. [Google Scholar]
- Gilbert, D.N.; Chambers, H.F.; Eliopoulos, G.M.; Saag, M.S. The Sanford Guide to Antimicrobial Therapy 2014, 44th ed.; Antimicrobial Therapy Inc: Sperryville, VA, USA, 2014. [Google Scholar]
- Tribble, D.R. Resistant pathogens as causes of traveller’s diarrhea globally and impact(s) on treatment failure and recommendations. J. Travel Med. 2017, 24, S6–S12. [Google Scholar] [CrossRef] [Green Version]
- Keusch, G.T.; Walker, C.F.; Das, J.K.; Horton, S.; Habte, D. Chapter 9. Diarrheal diseases. In Reproductive, Maternal, Newborn, and Child Health: Disease Control Priorities, 3rd ed.; Black, R.E., Ed.; The International Bank for Reconstruction and Development/The World Bank: Washington, DC, USA, 2016; pp. 163–185. [Google Scholar]
- Holloway, K.A.; Rosella, L.; Henry, D. The impact of WHO Essential Medicines policies on inappropriate use of antibiotics. PLoS ONE 2016, 11, e0152020. [Google Scholar] [CrossRef] [Green Version]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Boeckel, T.P.; Glennon, E.E.; Chen, D.; Gilbert, M.; Robinson, T.P.; Grenfell, B.T.; Levin, S.A.; Bonhoeffer, S.; Laxminarayan, R. Reducing antimicrobial use in food animals. Science 2017, 357, 1350–1352. [Google Scholar] [CrossRef] [Green Version]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palma, E.; Tilocca, B.; Roncada, P. Antimicrobial resistance in veterinary medicine: An overview. Int. J. Mol. Sci. 2020, 21, 1914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henriksson, P.J.G.; Troell, M.; Rico, A. Antimicrobial use in aquaculture: Some complementing facts. Proc. Natl. Acad. Sci. USA 2015, 112, E3317. [Google Scholar] [CrossRef] [Green Version]
- Santos, L.; Ramos, F. Antimicrobial resistance in aquaculture: Current knowledge and alternatives to tackle the problem. Int. J. Antimicrob. Agents 2018, 52, 135–143. [Google Scholar] [CrossRef]
- Schar, D.; Sommanustweechai, A.; Laxminarayan, R.; Tangcharoensathien, V. Surveillance of antimicrobial consumption in animal production sectors of low- and middle-income countries: Optimizing use and addressing antimicrobial resistance. PLoS Med. 2018, 15, e1002521. [Google Scholar] [CrossRef] [Green Version]
- Huijbers, P.M.C.; Blaak, H.; De Jong, M.C.M.; Graat, E.A.M.; Vandenbroucke-Grauls, C.M.J.E.; Husman, A.M.D.R.; Venbroucke-Grauls, C.M.J.E. Role of the environment in the transmission of antimicrobial resistance to humans: A review. Environ. Sci. Technol. 2015, 49, 11993–12004. [Google Scholar] [CrossRef]
- Richardson, E.J.; Bacigalupe, R.; Harrison, E.M.; Weinert, L.A.; Lycett, S.J.; Vrieling, M.; Robb, K.; Hoskisson, P.A.; Holden, M.T.G.; Feil, E.J.; et al. Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2018, 2, 1468–1478. [Google Scholar] [CrossRef]
- Banerji, A.; Jahne, M.; Herrmann, M.; Brinkman, N.; Keely, S. Bringing community ecology to bear on the issue of antimicrobial resistance. Front. Microbiol. 2019, 10, 2626. [Google Scholar] [CrossRef] [Green Version]
- Collignon, P.A.; McEwen, S. One Health—Its importance in helping to better control antimicrobial resistance. Trop. Med. Infect. Dis. 2019, 4, 22. [Google Scholar] [CrossRef] [Green Version]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance—The need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; United Kingdom Government, Wellcome Trust: London, UK, 2016. [Google Scholar]
- O’Neill, J. The Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; United Kingdom Government, Wellcome Trust: London, UK, 2014.
- Chopra, I.; Roberts, M.C. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. WHO Report on Surveillance of Antibiotic Consumption: 2016–2018 Early Implementation; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- World Health Organization. Critically Important Antimicrobials for Human Medicine, 5th Revision 2016; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Bryce, A.; Costelloe, C.; Hawcroft, C.; Wootton, M.; Hay, A.D. Faecal carriage of antibiotic resistant Escherichia coli in asymptomatic children and associations with primary care antibiotic prescribing: A systematic review and meta-analysis. BMC Infect. Dis. 2016, 16, 359. [Google Scholar] [CrossRef] [PubMed]
- Grossman, T.H. Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Patel, S.; Wencewicz, T.A.; Dantas, G. The tetracycline destructases: A novel family of tetracycline-inactivating enzymes. Chem. Biol. 2015, 22, 888–897. [Google Scholar] [CrossRef] [Green Version]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Mechanism of Resistance for Characterized Tet and Otr Genes. Available online: https://faculty.washington.edu/marilynr/tetweb1.pdf (accessed on 3 August 2020).
- You, Y.; Hilpert, M.; Ward, M.J. Identification of Tet45, a tetracycline efflux pump, from a poultry-litter-exposed soil isolate and persistence of tet(45) in the soil. J. Antimicrob. Chemother. 2013, 68, 1962–1969. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. The Global View of Campylobacteriosis: Report of an Expert Consultation; Utrecht, The Netherlands, 9–11 July 2012; World Health Organization: Geneva, Switzerland, 2013. [Google Scholar]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global epidemiology of campylobacter infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef] [Green Version]
- Taitt, C.R.; Leski, T.; Stockelman, M.G.; Craft, D.W.; Zurawski, D.V.; Kirkup, J.B.C.; Vora, G.J. Antimicrobial resistance determinants in acinetobacter baumannii isolates taken from military treatment facilities. Antimicrob. Agents Chemother. 2013, 58, 767–781. [Google Scholar] [CrossRef] [Green Version]
- Abril, C.; Brodard, I.; Perreten, V. Two novel antibiotic resistance genes, tet(44) and ant(6)-Ib, are located within a transferable pathogenicity island in Campylobacter fetus subsp. fetus. Antimicrob. Agents Chemother. 2010, 54, 3052–3055. [Google Scholar] [CrossRef] [Green Version]
- Charvalos, E.; Tselentis, Y.; Hamzehpour, M.M.; Kohler, T.; Pechere, J.C. Evidence for an efflux pump in multidrug-resistant Campylobacter jejuni. Antimicrob. Agents Chemother. 1995, 39, 2019–2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, M.C.; Schwarz, S.; Aarts, H.J.M. Erratum: Acquired antibiotic resistance genes: An overview. Front. Microbiol. 2012, 3, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taitt, C.R.; Leski, T.; Chen, A.; Berk, K.L.; Dorsey, R.W.; Gregory, M.J.; Sozhamannan, S.; Frey, K.G.; Dutt, D.L.; Vora, G.J. A survey of antimicrobial resistance determinants in category A Select Agents, exempt strains, and near-neighbor species. Int. J. Mol. Sci. 2020, 21, 1669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taitt, C.R.; Leski, T.A.; Colston, S.M.; Bernal, M.; Canal, E.; Regeimbal, J.; Rios, P.; Vora, G.J. A comparison of methods for DNA preparation prior to microarray analysis. Anal. Biochem. 2019, 585, 113405. [Google Scholar] [CrossRef]
- Nirdnoy, W.; Mason, C.J.; Guerry, P. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob. Agents Chemother. 2005, 49, 2454–2459. [Google Scholar] [CrossRef] [Green Version]
- Qin, S.; Wang, Y.; Zhang, Q.; Chen, X.; Shen, Z.; Deng, F.; Wu, C.; Shen, J. Identification of a novel genomic island conferring resistance to multiple aminoglycoside antibiotics in Campylobacter coli. Antimicrob. Agents Chemother. 2012, 56, 5332–5339. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, M.; Deng, F.; Shen, Z.; Wu, C.; Zhang, J.; Zhang, Q.; Shen, J. Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob. Agents Chemother. 2014, 58, 5405–5412. [Google Scholar] [CrossRef] [Green Version]
- Igwaran, A.; Okoh, A.I. Campylobacteriosis agents in meat carcasses collected from two distinct municipalities in the Eastern Cape Province, South Africa. Foods 2020, 9, 203. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases; 2018, World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Chesneau, O.; Tsvetkova, K.; Courvalin, P. Resistance phenotypes conferred by macrolide phosphotransferases. FEMS Microbiol. Lett. 2007, 269, 317–322. [Google Scholar] [CrossRef]
- Obiesie, N.; Flahart, R.; Hansen, G.; Sexton, J.; Pursell, C.; Sugg, T.J.; Thoroughman, D.A.; Humbaugh, K.E.; Zhu, B.P.; Hinkle, C.J.; et al. Outbreaks of multidrug-resistant Shigella sonnei gastroenteritis associated with day care centers—Kansas, Kentucky, and Missouri, 2005. Morb. Mortal. Weekly Rep. 2006, 55, 1068–1071. [Google Scholar]
- World Health Organization. Guidelines for the Control of Shigellosis, Including Epidemics Due to Shigella Dysenteriae Type 1; World Health Organization: Geneva, Switzerland, 2005. [Google Scholar]
- Tauxe, R.V.; Cavanagh, T.R.; Cohen, M.L. Interspecies gene transfer in vivo producing an outbreak of multiply resistant shigellosis. J. Infect. Dis. 1989, 160, 1067–1070. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.C.P.; Woerther, P.L.; Bouvet, M.; Andremont, A.; Leclercq, R.; Canu, A. Escherichia coli as reservoir for macrolide resistance genes. Emerg. Infect. Dis. 2009, 15, 1648–1650. [Google Scholar] [CrossRef] [PubMed]
- Darton, T.C.; Tuyen, H.T.; The, H.C.; Newton, P.N.; Dance, D.A.; Phetsouvanh, R.; Davong, V.; Campbell, J.I.; Hoang, N.V.M.; Thwaites, G.E.; et al. Azithromycin resistance in Shigella spp. in Southeast Asia. Antimicrob. Agents Chemother. 2018, 62, e01748-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taitt, C.R.; Leski, T.; Erwin, D.P.; Odundo, E.A.; Kipkemoi, N.C.; Ndonye, J.N.; Kirera, R.K.; Ombogo, A.N.; Walson, J.L.; Pavlinac, P.B.; et al. Antimicrobial resistance of Klebsiella pneumoniae stool isolates circulating in Kenya. PLoS ONE 2017, 12, e0178880. [Google Scholar] [CrossRef] [Green Version]
- Amos, G.C.A.; Ploumakis, S.; Zhang, L.; Hawkey, P.M.; Gaze, W.H.; Wellington, E.M.H. The widespread dissemination of integrons throughout bacterial communities in a riverine system. ISME J. 2018, 12, 681–691. [Google Scholar] [CrossRef] [Green Version]
- Kiiru, J.; Butaye, P.; Goddeeris, B.; Kariuki, S. Analysis for prevalence and physical linkages amongst integrons, ISEcp1, ISCR1, Tn21 and Tn7 encountered in Escherichia coli strains from hospitalized and non-hospitalized patients in Kenya during a 19-year period (1992–2011). BMC Microbiol. 2013, 13, 109. [Google Scholar] [CrossRef] [Green Version]
- Handford, C.; Stang, C.; Raivio, T.; Dennis, J. The contribution of small cryptic plasmids to the antibiotic resistance of enteropathogenic Escherichia coli E2348/69. Can. J. Microbiol. 2009, 55, 1229–1239. [Google Scholar] [CrossRef]
- Yau, S.; Liu, X.; Djordjevic, S.P.; Hall, R. RSF1010-like plasmids in Australian Salmonella enterica serovar typhimurium and origin of their sul2-strA-strB antibiotic resistance gene cluster. Microb. Drug Resist. 2010, 16, 249–252. [Google Scholar] [CrossRef]
- Kobayashi, N.; Nishino, K.; Yamaguchi, A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 2001, 183, 5639–5644. [Google Scholar] [CrossRef] [Green Version]
- Khalil, A.I.; Troeger, C.; Blacker, B.F.; Rao, P.C.; Brown, A.E.; Atherly, D.; Brewer, T.G.; Engmann, C.M.; Houpt, E.R.; Kang, G.; et al. Morbidity and mortality due to Shigella and enterotoxigenic Escherichia coli diarrhoea: The Global Burden of Disease Study 1990–2016. Lancet Infect. Dis. 2018, 18, 1229–1240. [Google Scholar] [CrossRef] [Green Version]
- Chattaway, M.A.; Schaefer, U.; Tewolde, R.; Dallman, T.J.; Jenkins, C. Identification of Escherichia coli and Shigella species from whole-genome sequences. J. Clin. Microbiol. 2016, 55, 616–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Njamkepo, E.; Fawal, N.; Tran-Dien, A.; Hawkey, J.; Strockbine, N.; Jenkins, C.; Talukder, K.A.; Bercion, R.; Kuleshov, K.; Kolínská, R.; et al. Global phylogeography and evolutionary history of Shigella dysenteriae type 1. Nat. Microbiol. 2016, 1, 16027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.K.; Speelman, P.; Butler, T.; Nath, S.; Rahman, H.; Stoll, B.J. Diarrhea associated with typhoid fever. J. Infect. Dis. 1985, 151, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Parry, C.M.; Hien, T.T.; Dougan, G.; White, N.J.; Farrar, J.J. Typhoid fever. N. Engl. J. Med. 2002, 347, 1770–1782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mushi, M.F.; Mshana, S.E.; Imirzalioglu, C.; Bwanga, F. Carbapenemase genes among multidrug resistant gram negative clinical isolates from a tertiary hospital in Mwanza, Tanzania. BioMed Res. Int. 2014, 2014, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Okoche, D.; Asiimwe, B.B.; Katabazi, F.A.; Kato, L.; Najjuka, C.F. Prevalence and characterization of carbapenem-resistant enterobacteriaceae isolated from Mulago National Referral Hospital, Uganda. PLoS ONE 2015, 10, e0135745. [Google Scholar] [CrossRef]
- Ssekatawa, K.; Byarugaba, D.K.; Wampande, E.; Francis, E. A systematic review: The current status of carbapenem resistance in East Africa. BMC Res. Notes 2018, 11, 629. [Google Scholar] [CrossRef] [Green Version]
- Ampaire, L.M.; Katawera, V.; Nyehangane, D.; Boum, Y.; Bazira, J. Epidemiology of carbapenem resistance among multi-drug resistant Enterobacteriaceae in Uganda. Br. Microbiol. Res. J. 2015, 8, 418–423. [Google Scholar] [CrossRef] [Green Version]
- Antunes, P.; Machado, J.; Peixe, L. Characterization of antimicrobial resistance and class 1 and 2 integrons in Salmonella enterica isolates from different sources in Portugal. J. Antimicrob. Chemother. 2006, 58, 297–304. [Google Scholar] [CrossRef]
- Ploy, M.C.; Chainier, D.; Thi, N.H.T.; Poilane, I.; Cruaud, P.; Denis, F.; Collignon, A.; Lambert, T. Integron-associated antibiotic resistance in Salmonella enterica serovar Typhi from Asia. Antimicrob. Agents Chemother. 2003, 47, 1427–1429. [Google Scholar] [CrossRef] [Green Version]
- Van Essen-Zandbergen, A.; Smith, H.; Veldman, K.; Mevius, D. Occurrence and characteristics of class 1, 2 and 3 integrons in Escherichia coli, Salmonella and Campylobacter spp. in The Netherlands. J. Antimicrob. Chemother. 2007, 59, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Boyd, D.; Peters, G.A.; Cloeckaert, A.; Sidi-Boumedine, K.; Chaslus-Dancla, E.; Imberechts, H.; Mulvey, M.R. Complete nucleotide sequence of a 43-kilobase genomic island associated with the Multidrug Resistance Region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 2001, 183, 5725–5732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Hello, S.; Hendriksen, R.S.; Doublet, B.; Fisher, I.; Nielsen, E.M.; Whichard, J.M.; Bouchrif, B.; Fashae, K.; Granier, S.A.; Silva, N.J.D.; et al. International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J. Infect. Dis. 2011, 204, 675–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdulrahman, E.; El-Sherif, R.H. High rates of intestinal colonization with extended-spectrum lactamase-producing Enterobacteriaceae among healthy individuals. J. Investig. Med. 2011, 59, 1284–1286. [Google Scholar] [CrossRef]
- Abdallah, H.M.; Al Naiemi, N.; Reuland, E.A.; Wintermans, B.B.; Koek, A.; Abdelwahab, A.; Samy, A.; Abdelsalam, K.; Vandenbroucke-Grauls, C.M.J.E. Fecal carriage of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae in Egyptian patients with community-onset gastrointestinal complaints: A hospital -based cross-sectional study. Antimicrob. Resist. Infect. Control 2017, 6, 62. [Google Scholar] [CrossRef]
- Borg, M.; Zarb, P.; Ferech, M.; Goossens, H.; on behalf of the ARMed Project Group. Antibiotic consumption in southern and eastern Mediterranean hospitals: Results from the ARMed project. J. Antimicrob. Chemother. 2008, 62, 830–836. [Google Scholar] [CrossRef] [Green Version]
- Center for Disease Dynamics. E.P. State of the World’s Antibiotics, 2015; CDDEP: Washington, DC, USA, 2015. [Google Scholar]
- Thompson, C.N.; Phan, M.V.T.; Hoang, N.V.M.; Van Minh, P.; Vinh, N.T.; Thuy, C.T.; Nga, T.T.T.; Rabaa, M.A.; Duy, P.T.; Dung, T.T.N.; et al. A prospective multi-center observational study of children hospitalized with diarrhea in Ho Chi Minh City, Vietnam. Am. J. Trop. Med. Hyg. 2015, 92, 1045–1052. [Google Scholar] [CrossRef]
- Challacombe, J.F.; Altherr, M.R.; Xie, G.; Bhotika, S.S.; Brown, N.; Bruce, D.; Campbell, C.S.; Campbell, M.L.; Chen, J.; Chertkov, O.; et al. The complete genome sequence of Bacillus thuringiensis Al Hakam. J. Bacteriol. 2007, 189, 3680–3681. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.Q.; Guo, H.; Wu, J.H. Effects of target length on the hybridization efficiency and specificity of rRNA-based oligonucleotide microarrays. Appl. Environ. Microbiol. 2006, 73, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Taitt, C.R.; Lêski, T.; Heang, V.; Ford, G.W.; Prouty, M.G.; Newell, S.W.; Vora, G.J. Antimicrobial resistance genotypes and phenotypes from multidrug-resistant bacterial wound infection isolates in Cambodia. J. Glob. Antimicrob. Resist. 2015, 3, 198–204. [Google Scholar] [CrossRef] [Green Version]
- Kariuki, S.; Okoro, C.K.; Kiiru, J.; Njoroge, S.; Omuse, G.; Langridge, G.C.; Kingsley, R.A.; Dougan, G.; Revathi, G. Ceftriaxone-resistant Salmonella enterica serotype Typhimurium sequence type 313 from Kenyan patients is associated with the blaCTX-M-15 gene on a novel IncHI2 plasmid. Antimicrob. Agents Chemother. 2015, 59, 3133–3139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connor, T.R.; Barker, C.; Baker, K.S.; Weill, F.-X.; Talukder, K.A.; Smith, A.M.; Baker, S.; Gouali, M.; Thanh, D.P.; Azmi, I.J.; et al. Species-wide whole genome sequencing reveals historical global spread and recent local persistence in Shigella flexneri. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Global Antibiotic Resistance Partnership-Kenya Working Group. Situational Analysis and Recommendations—Antibiotic Use and Resistance in Kenya; Center for Disease Dynamics, Economics & Policy: Washington, DC, USA, 2011. [Google Scholar]
- Henson, S.; Boinett, C.J.; Ellington, M.J.; Kagia, N.; Mwarumba, S.; Nyongesa, S.; Mturi, N.; Kariuki, S.; Scott, J.A.G.; Thomson, N.R.; et al. Molecular epidemiology of Klebsiella pneumoniae invasive infections over a decade at Kilifi County Hospital in Kenya. Int. J. Med. Microbiol. 2017, 307, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Manenzhe, R.I.; Zar, H.J.; Nicol, M.P.; Kaba, M. The spread of carbapenemase-producing bacteria in Africa: A systematic review. J. Antimicrob. Chemother. 2014, 70, 23–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamprecht, A.; Sommer, J.; Willmann, M.; Brender, C.; Stelzer, Y.; Krause, F.F.; Tsvetkov, T.; Wild, F.; Riedel-Christ, S.; Kutschenreuter, J.; et al. Pathogenicity of clinical OXA-48 isolates and impact of the OXA-48 IncL plasmid on virulence and bacterial fitness. Front. Microbiol. 2019, 10, 2509. [Google Scholar] [CrossRef]
- Di Luca, M.C.; Johnsen, P.; Samuelsen, Ø. Acquisition of Clinical Carbapenemase-Encoding Plasmids Confers Limited Fitness Cost but Results in Variable Plasmid Stability in Clinical Escherichia Coli Strains, in European Congress of Clinical Microbiology and Infectious Diseases, 2018; ECCMID: Madrid, Spain, 2018. [Google Scholar]
- Mwansa, B.J. The Effect of Sub-Lethal Concentration of Ciprofloxacin on the Transfer of Multidrug Resistance Plasmids, Fitness Costs on the Host and the Stability of the Newly Acquired Plasmids; University of Tromso: Tromso, Norway, 2014. [Google Scholar]
- Wein, T.; Hülter, N.F.; Mizrahi, I.; Dagan, T. Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat. Commun. 2019, 10, 2595. [Google Scholar] [CrossRef] [Green Version]
- Carroll, A.C.; Wong, A. Plasmid persistence: Costs, benefits, and the plasmid paradox. Can. J. Microbiol. 2018, 64, 293–304. [Google Scholar] [CrossRef]
- Porse, A.; Schønning, K.; Munck, C.; Sommer, M.O. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol. Biol. Evol. 2016, 33, 2860–2873. [Google Scholar] [CrossRef]
- Ludden, C.M.; Raven, K.E.; Jamrozy, D.; Gouliouris, T.; Blane, B.; Coll, F.; De Goffau, M.; Naydenova, P.; Horner, C.; Hernandez-Garcia, J.; et al. One health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. mBio 2019, 10, e02693-18. [Google Scholar] [CrossRef] [Green Version]
- Snitkin, E.S.; Zelazny, A.M.; Thomas, P.J.; Stock, F.; Henderson, D.K.; Palmore, T.N.; Segre, J.A.; NISC Comparative Sequencing Program Group. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 2012, 4, 148ra116. [Google Scholar] [CrossRef] [Green Version]
- Tyson, G.H.; McDermott, P.F.; Li, C.; Chen, Y.; Tadesse, D.A.; Mukherjee, S.; Bodeis-Jones, S.; Kabera, C.; Gaines, S.A.; Loneragan, G.H.; et al. WGS accurately predicts antimicrobial resistance in Escherichia coli. J. Antimicrob. Chemother. 2015, 70, 2763–2769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leski, T.A.; Stockelman, M.G.; Bangura, U.; Chae, D.; Ansumana, R.; Stenger, D.A.; Vora, G.J.; Taitt, C.R. Prevalence of quinolone resistance in Enterobacteriaceae from Sierra Leone and the detection of qnrB pseudogenes and modified LexA binding sites. Antimicrob. Agents Chemother. 2016, 60, 6920–6923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamowicz, E.M.; Flynn, J.; Hunter, R.C.; Harcombe, W.R. Cross-feeding modulates antibiotic tolerance in bacterial communities. ISME J. 2018, 12, 2723–2735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamowicz, E.M.; Muza, M.; Chacón, J.M.; Harcombe, W.R. Cross-feeding modulates the rate and mechanism of antibiotic resistance evolution in a model microbial community of Escherichia coli and Salmonella enterica. PLoS Pathog. 2020, 16, e1008700. [Google Scholar] [CrossRef] [PubMed]
- Olsen, I. Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Islas-Espinoza, M.; Reid, B.J.; Wexler, M.; Bond, P.L. Soil bacterial consortia and previous exposure enhance the biodegradation of sulfonamides from pig manure. Microb. Ecol. 2012, 64, 140–151. [Google Scholar] [CrossRef]
- Sayed, A.M.; Behiry, I.K.; Elsherief, R.H.; Elsir, S.A. Detection of carbapenemase-producing Enterobacteriaceae in rectal surveillance cultures in non-hospitalized patients. J. Anal. Sci. Technol. 2017, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Tran, D.M.; Larsson, M.; Olson, L.; Hoang, N.T.B.; Le, N.K.; Khu, D.T.K.; Nguyen, H.D.; Vu, T.V.; Trinh, T.H.; Le, T.Q.; et al. High prevalence of colonisation with carbapenem-resistant Enterobacteriaceae among patients admitted to Vietnamese hospitals: Risk factors and burden of disease. J. Infect. 2019, 79, 115–122. [Google Scholar] [CrossRef] [Green Version]
- Riddle, M.S.; Putnam, S.D.; Tribble, D.R.; Sanders, J.W. Incidence, etiology, and impact of diarrhea among long-term travelers (US military and similar populations): A systematic review. Am. J. Trop. Med. Hyg. 2006, 74, 891–900. [Google Scholar] [CrossRef] [Green Version]
- Lavilla, S.; González-López, J.J.; Sabaté, M.; Larrosa, M.N.; García-Fernández, A.; Bartolomé, R.M.; Carattoli, A.; Prats, G. Prevalence of qnr genes among extended-spectrum β-lactamase-producing enterobacterial isolates in Barcelona, Spain. J. Antimicrob. Chemother. 2007, 61, 291–295. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Chen, Y.; Wu, Z.; Zhao, Z.; Wu, J.; Cao, Y. Prevalence of plasmid-mediated quinolone resistance genes among Escherichia coli in the gut of healthy people in Fuzhou, China. Ann. Lab. Med. 2018, 38, 384–386. [Google Scholar] [CrossRef] [PubMed]
- Riveros, M.; Riccobono, E.; Durand, D.; Mosquito, S.; Ruiz, J.; Rossolini, G.M.; Ochoa, T.J.; Pallecchi, L. Plasmid-mediated quinolone resistance genes in enteroaggregative Escherichia coli from infants in Lima, Peru. Int. J. Antimicrob. Agents 2012, 39, 540–542. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: Quinolone resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, P.R.; Baron, E.J.; Jorgensen, J.H.; Pfaller, M.A.; Yolken, R.H. Manual of Clinical Microbiology, 8th ed.; American Society for Microbiology: Washington, DC, USA, 2003. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Nineteenth Informational Supplement M100-S19; CLSI: Wayne, PA, USA, 2009. [Google Scholar]
- Alexopoulou, K.; Foka, A.; Petinaki, E.; Jelastopulo, E.; Dimitracoupoulos, G.; Spiliopoulou, I. Comparison of two commercial methods with PCR restriction fragment polymorphism of the tuf gene in the identification of coagulase-hegative staphylococci. Lett. Appl. Microbiol. 2006, 43, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Lêski, T.; Vora, G.J.; Barrows, B.R.; Pimentel, G.; House, B.L.; Nicklasson, M.; Wasfy, M.; Abdel-Maksoud, M.; Taitt, C.R. Molecular characterization of multidrug resistant hospital isolates using the antimicrobial resistance determinant microarray. PLoS ONE 2013, 8, e69507. [Google Scholar] [CrossRef] [Green Version]
- Koeleman, J.G.M.; Stoof, J.; Van Der Bijl, M.W.; Vandenbroucke-Grauls, C.M.J.E.; Savelkoul, P.H. Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J. Clin. Microbiol. 2001, 39, 8–13. [Google Scholar] [CrossRef] [Green Version]




| By Genus | ||||||||
| No. of Antimicrobial Classes | Campylobacter | E. coli | Salmonella | Shigella | ||||
| 0 | 9 | 0 | 0 | 0 | ||||
| 1 | 74 | 0 | 3 | 4 | ||||
| 2 | 9 | 3 | 6 | 7 | ||||
| 3 | 4 | 17 | 13 | 11 | ||||
| 4 | 4 | 13 | 13 | 4 | ||||
| 5 | 0 | 10 | 25 | 20 | ||||
| 6 | 0 | 13 | 31 | 4 | ||||
| 7 | 0 | 27 | 9 | 31 | ||||
| 8 | 0 | 17 | 0 | 16 | ||||
| 9 | 0 | 0 | 0 | 2 | ||||
| % potentially resistant to: | ||||||||
| ≥3 classes | 8 | 97 | 91 | 89 | ||||
| ≥6 classes | 0 | 57 | 40 | 53 | ||||
| No. of isolates tested | 23 | 30 | 32 | 45 | ||||
| By geographic location | ||||||||
| No. of Antimicrobial Classes | USA | Cambodia | Egypt | Peru | Kenya | |||
| 0 | 0 | 0 | 0 | 8 | 0 | |||
| 1 | 15 | 30 | 16 | 17 | 0 | |||
| 2 | 7 | 13 | 6 | 4 | 0 | |||
| 3 | 19 | 17 | 0 | 8 | 17 | |||
| 4 | 15 | 13 | 3 | 4 | 4 | |||
| 5 | 15 | 4 | 9 | 25 | 33 | |||
| 6 | 7 | 4 | 16 | 13 | 17 | |||
| 7 | 19 | 17 | 22 | 13 | 25 | |||
| 8 | 4 | 0 | 28 | 4 | 4 | |||
| 9 | 0 | 0 | 0 | 4 | 0 | |||
| % potentially resistant to: | ||||||||
| ≥3 classes | 78 | 57 | 78 | 71 | 100 | |||
| ≥6 classes | 30 | 21 | 66 | 34 | 46 | |||
| No. of isolates tested | 27 | 23 | 32 | 24 | 24 | |||
| ARD | USA (n = 5) | Cambodia (n = 4) | Egypt (n = 6) | Peru (n = 7) | Kenya (n = 1) |
|---|---|---|---|---|---|
| aadE | nd * | nd | nd | 14% | 100% |
| aphA3 | 20% | nd | nd | 14% | 100% |
| erm(B) | nd | nd | nd | nd | 100% |
| tet(M) | nd | nd | nd | nd | 100% |
| tet(O)/tet(32) | 100% | 100% | 100% | 71% | nd |
| sat4 | nd | nd | 17% | 14% | 100% |
| ARD | USA (n = 7) | Cambodia (n = 4) | Egypt (n = 8) | Peru (n = 3) | Kenya (n = 8) |
|---|---|---|---|---|---|
| blaLEN | nd * | nd | 13% | nd | nd |
| blaOXA-1 family | nd | nd | 13% | nd | nd |
| blaOXA-9 family | nd | nd | 13% | nd | nd |
| blaSHV family | nd | nd | 25% | nd | nd |
| blaTEM family | 14% | 100% | 88% | 67% | 63% |
| blaCTX-M-1 family | nd | nd | 38% | nd | nd |
| blaCTX-M-9 family | nd | nd | 38% | nd | nd |
| blaNDM | nd | nd | 13% | nd | nd |
| aac(6)-Ib | nd | nd | 13% | nd | nd |
| aadA1/A2 | nd | nd | 63% | 67% | nd |
| aphA1 | nd | nd | 25% | nd | nd |
| aphA4 | 14% | nd | nd | nd | nd |
| strA | 57% | 75% | 75% | 33% | 88% |
| strB | 57% | 75% | 75% | 33% | 88% |
| aphA6 | nd | nd | 13% | nd | nd |
| mac(A) | 86% | 100% | 88% | 100% | 38% |
| mac(B) | 86% | 25% | 88% | 100% | 38% |
| mph(A)/mph(K) | 14% | 25% | 50% | nd | 25% |
| tet(A) | 29% | 25% | 50% | nd | 25% |
| tet(B) | 43% | 75% | 50% | 33% | 50% |
| tet(C) | 29% | nd | nd | nd | nd |
| tet(D) | nd | nd | 3% | nd | nd |
| catA1/cat4 | nd | 25% | 25% | 33% | 25% |
| cmr | 100% | 100% | 100% | 100% | 38% |
| qnrS | nd | 25% | 13% | nd | nd |
| qacEΔ1 | 14% | nd | 13% | 33% | 13% |
| sat2 | nd | nd | 38% | 33% | nd |
| sul1 | 14% | nd | 25% | 33% | 13% |
| sul2 | 57% | 75% | 63% | 33% | 88% |
| dfrA1 | nd | nd | 50% | nd | nd |
| dfrA5 | nd | nd | nd | nd | 13% |
| dfrA7 | 14% | nd | nd | nd | 13% |
| dfrA8 | nd | 50% | nd | 33% | 25% |
| dfrA12 | nd | nd | 13% | nd | nd |
| dfrA14 | nd | nd | nd | nd | 63% |
| dfrA17 | 14% | nd | nd | nd | nd |
| dfrA19 | nd | nd | 25% | nd | nd |
| ARD | USA (n = 7) | Cambodia (n = 11) | Egypt (n = 12) | Peru (n = 6) | Kenya (n = 9) |
|---|---|---|---|---|---|
| blaLEN | nd * | nd | 8% | nd | nd |
| blaOXA-1 family | 29% | 9% | 67% | 67% | nd |
| blaSHV family | nd | nd | 8% | nd | nd |
| blaTEM family | 14% | 18% | 25% | 17% | 56% |
| blaCTX-M-1 family | nd | nd | 8% | nd | nd |
| blaCTX-M-9 family | nd | nd | 17% | nd | nd |
| aac(6)-Ib | nd | nd | nd | 17% | nd |
| aadA1/A2 | 57% | 18% | 100% | 67% | 44% |
| aadA4 | nd | nd | nd | 17% | nd |
| strA | 57% | 27% | 50% | 50% | 100% |
| strB | 57% | 36% | 50% | 50% | 100% |
| mac(A) | 86% | 18% | 100% | 100% | 33% |
| mac(B) | 86% | 9% | 83% | 100% | 33% |
| mph(A)/mph(K) | nd | nd | nd | 17% | 33% |
| tet(A) | 57% | 18% | 33% | nd | 78% |
| tet(B) | 29% | 73% | 67% | 100% | 11% |
| arr | nd | nd | nd | 17% | nd |
| catA1/cat4 | 14% | 18% | 58% | 67% | nd |
| cmr | 100% | 73% | 100% | 100% | 44% |
| qnrS | nd | nd | 8% | nd | 11% |
| qacEΔ1 | 14% | nd | 8% | 17% | 11% |
| sat2 | 43% | 18% | 67% | 17% | 44% |
| sul1 | 14% | nd | nd | 14% | 11% |
| sul2 | 57% | 27% | 75% | 67% | 100% |
| dfrA1 | 43% | 18% | 75% | 17% | 44% |
| dfrA5 | nd | nd | nd | nd | 11% |
| dfrA7 | 14% | nd | nd | nd | 11% |
| dfrA8 | nd | nd | nd | 14% | 11% |
| dfrA14 | nd | 9% | 8% | 33% | 11% |
| dfrA17 | nd | nd | nd | 17% | nd |
| ARD | USA (n = 8) | Cambodia (n = 4) | Egypt (n = 6) | Peru (n = 8) | Kenya (n = 6) |
|---|---|---|---|---|---|
| blaCMY/LAT | 25% | nd * | nd | nd | nd |
| blaLEN | nd | nd | 33% | nd | nd |
| blaOXA-48 family | nd | nd | nd | nd | 17% |
| blaPSE/CARB | 13% | nd | 17% | nd | nd |
| blaSHV family | nd | nd | 50% | nd | nd |
| blaTEM family | 50% | 50% | 83% | 25% | 83% |
| blaCTX-M-1 family | nd | 25% | 17% | nd | nd |
| blaCTX-M-9 family | nd | nd | 33% | nd | nd |
| aac(3)-Id | nd | nd | 50% | nd | nd |
| aac(3)-III | nd | nd | nd | 13% | nd |
| aadA1/A2 | 50% | nd | 50% | 75% | 50% |
| aadA7 | nd | nd | 50% | nd | nd |
| aphA1 | 13% | nd | 50% | 63% | nd |
| strA | 63% | 75% | 50% | 25% | 83% |
| strB | 63% | 75% | 67% | 25% | 100% |
| aphA6 | 13% | nd | 33% | nd | nd |
| rmtD | 13% | nd | nd | nd | 17% |
| ere(A2) | nd | nd | 33% | nd | nd |
| mph(A)/mph(K) | nd | nd | 17% | nd | 17% |
| tet(A) | 38% | 50% | 33% | 88% | 17% |
| tet(B) | 50% | 50% | nd | nd | 17% |
| tet(C) | 13% | nd | nd | nd | nd |
| tet(D) | nd | nd | 17% | 13% | nd |
| tet(G) | 13% | nd | 17% | nd | nd |
| catA1/cat4 | 25% | nd | nd | nd | 33% |
| floR | 25% | nd | 17% | 13% | 17% |
| cmr | nd | nd | nd | nd | 17% |
| qnrS | nd | 25% | nd | nd | nd |
| qacEΔ1 | 38% | nd | 83% | 75% | 50% |
| sul1 | 38% | nd | 83% | 75% | 50% |
| sul2 | 50% | 75% | 33% | 13% | 100% |
| sul3 | nd | nd | nd | 13% | nd |
| dfrA1 | nd | nd | nd | 13% | 33% |
| dfrA7 | 25% | nd | nd | nd | 17% |
| dfrA8 | nd | nd | nd | nd | 17% |
| dfrA12 | nd | nd | 17% | 13% | nd |
| dfrA14 | nd | nd | nd | 63% | 50% |
| dfrA19 | nd | nd | 19% | nd | nd |
| Antimicrobial Class | Geographic Location | ||||
|---|---|---|---|---|---|
| USA | Cambodia | Egypt | Peru | Kenya | |
| β-Lactams | 50.0 | 42.1 | 88.5 | 47.1 | 65.2 |
| Aminoglycosides | 77.3 | 52.6 | 100.0 | 88.2 | 95.7 |
| Macrolides | 54.5 | 31.6 | 80.8 | 52.9 | 47.8 |
| Tetracyclines | 95.4 | 94.7 | 92.3 | 88.3 | 70.0 |
| Ansamycins | nd ** | nd | nd | 5.9 | nd |
| Phenicols | 81.8 | 63.2 | 80.8 | 64.7 | 47.8 |
| Quinolones | nd | 10.5 | 7.7 | nd | 4.3 |
| Quaternary amines | 22.7 | nd | 26.9 | 47.1 | 21.7 |
| Streptothricins | 13.6 | 10.5 | 42.3 | 1.8 | 17.4 |
| Sulfonamides | 63.6 | 47.4 | 73.1 | 82.4 | 95.7 |
| Trimethoprim | 31.8 | 26.3 | 69.2 | 70.6 | 78.3 |
| Class 1 integron * | 18.2 | n/a *** | 19.2 | 47.1 | 17.4 |
| Class 2 integron * | 9.1 | n/a | 34.7 | 5.9 | 13.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taitt, C.R.; Leski, T.A.; Prouty, M.G.; Ford, G.W.; Heang, V.; House, B.L.; Levin, S.Y.; Curry, J.A.; Mansour, A.; Mohammady, H.E.; et al. Tracking Antimicrobial Resistance Determinants in Diarrheal Pathogens: A Cross-Institutional Pilot Study. Int. J. Mol. Sci. 2020, 21, 5928. https://doi.org/10.3390/ijms21165928
Taitt CR, Leski TA, Prouty MG, Ford GW, Heang V, House BL, Levin SY, Curry JA, Mansour A, Mohammady HE, et al. Tracking Antimicrobial Resistance Determinants in Diarrheal Pathogens: A Cross-Institutional Pilot Study. International Journal of Molecular Sciences. 2020; 21(16):5928. https://doi.org/10.3390/ijms21165928
Chicago/Turabian StyleTaitt, Chris R., Tomasz A. Leski, Michael G. Prouty, Gavin W. Ford, Vireak Heang, Brent L. House, Samuel Y. Levin, Jennifer A. Curry, Adel Mansour, Hanan El Mohammady, and et al. 2020. "Tracking Antimicrobial Resistance Determinants in Diarrheal Pathogens: A Cross-Institutional Pilot Study" International Journal of Molecular Sciences 21, no. 16: 5928. https://doi.org/10.3390/ijms21165928
APA StyleTaitt, C. R., Leski, T. A., Prouty, M. G., Ford, G. W., Heang, V., House, B. L., Levin, S. Y., Curry, J. A., Mansour, A., Mohammady, H. E., Wasfy, M., Tilley, D. H., Gregory, M. J., Kasper, M. R., Regeimbal, J., Rios, P., Pimentel, G., Danboise, B. A., Hulseberg, C. E., ... Vora, G. J. (2020). Tracking Antimicrobial Resistance Determinants in Diarrheal Pathogens: A Cross-Institutional Pilot Study. International Journal of Molecular Sciences, 21(16), 5928. https://doi.org/10.3390/ijms21165928