New Bacteriocins from Lacticaseibacillus paracasei CNCM I-5369 Adsorbed on Alginate Nanoparticles Are Very Active against Escherichia coli
Abstract
:1. Introduction
2. Results and Discussion
2.1. Peptide Mapping of the E20 Fraction
2.2. Characterization of Alginate Nanoparticles and Alginate Nanoparticles Loaded with E20
2.3. E20 Association with Alginate Nanoparticles’ Anti-E. coli Activity at Different pH Values
2.4. Determination of Minimal Inhibitory Concentration (MIC) against a Panel of E. coli Strains and Development of a Nano-Antibiotic Formulation
2.5. Antibacterial Activity
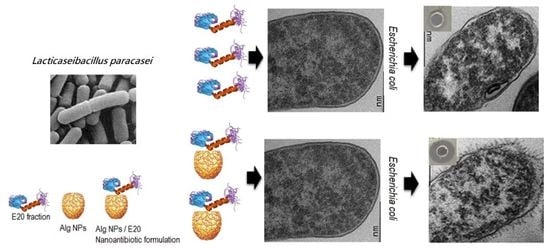
2.6. E20 and E20 Adsorbed on Alginate Nanoparticles Modify E. coli Cell Morphology
2.7. Nano-Antibiotic Formulation Alg NPs/E20 Is Not Toxic for Human (HT29) and Animal (IPEC-1) Cell Lines
3. Materials and Methods
3.1. Strains
3.2. Preparation of E20 Fraction and Its Analysis by Mass Spectrometry
3.3. Preparation and Characterization of Alginate Nanoparticles and Loading of the Active Fraction E20
3.4. High-Performance Liquid Chromatography (HPLC)
3.5. Scanning Electron Microscopy (SEM)
3.6. Minimal Inhibitory Concentration (MIC)
3.7. Time-Killing Curves
3.8. Transmission Electron Microscopy (TEM)
3.9. Proteins Quantification
3.10. Cytotoxicity Assay
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Golkar, Z.; Bagasra, O.; Pace, D.G. Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries 2014, 8, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2018, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Burnham, J.P.; Olsen, M.A.; Kollef, M.H. Re-estimating annual deaths due to multidrug-resistant organism infections. Infect. Control Hosp. Epidemiol. 2019, 40, 112–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CDC Centers for Disease Control and Prevention, Office of Infectious Disease Antibiotic Resistance Threats in the United States. 2013. Available online: http://www.cdc.gov/drugresistance/threat-report-2013 (accessed on 5 July 2019).
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [Green Version]
- Kempf, I.; Fleury, M.A.; Drider, D.; Bruneau, M.; Sanders, P.; Chauvin, C.; Madec, J.Y.; Jouy, E. What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? Int. J. Antimicrob. Agents 2013, 42, 379–383. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Nguyen, H.M.; Nguyen, C.V.; Nguyen, T.V.; Nguyen, M.T.; Thai, H.Q.; Ho, M.H.; Thwaites, G.; Ngo, H.T.; Baker, S.; et al. Use of Colistin and Other Critical Antimicrobials on Pig and Chicken Farms in Southern Vietnam and Its Association with Resistance in Commensal Escherichia coli Bacteria. Appl. Environ. Microbiol. 2016, 82, 3727–3735. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Irrgang, A.; Roschanski, N.; Tenhagen, B.A.; Grobbel, M.; Skladnikiewicz-Ziemer, T.; Thomas, K.; Roesler, U.; Käsbohrer, A. Prevalence of mcr-1 in E. coli from livestock and food in Germany, 2010–2015. PLoS ONE. 2016, 11, e0159863. [Google Scholar] [CrossRef]
- Ye, H.; Li, Y.; Li, Z.; Gao, R.; Zhang, H.; Wen, R.; Gao, G.F.; Hu, Q.; Feng, Y. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio 2016, 7, e00177-16. [Google Scholar] [CrossRef] [Green Version]
- Chiou, C.S.; Chen, Y.T.; Wang, Y.W.; Liu, Y.Y.; Kuo, H.C.; Tu, Y.H.; Lin, A.C.; Liao, Y.S.; Hong, Y.P. Dissemination of mcr-1-carrying plasmids among colistin-resistant Salmonella strains from humans and food-producing animals in Taiwan. Antimicrob. Agents Chemother. 2017, 61, e00338. [Google Scholar] [CrossRef] [Green Version]
- Falgenhauer, L.; Waezsada, S.E.; Gwozdzinski, K.; Ghosh, H.; Doijad, S.; Bunk, B.; Spröer, C.; Imirzalioglu, C.; Seifert, H.; Irrgang, A.; et al. Chromosomal Locations of mcr-1 and bla CTX-M-15 in Fluoroquinolone-Resistant Escherichia coli ST410. Emerg. Infect. Dis. 2016, 22, 1689–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalmolin, T.V.; de Lima-Morales, D.; Barth, A.L. Plasmid-mediated Colistin Resistance: What Do We Know? J. Infect. 2018, 1, 16–22. [Google Scholar] [CrossRef]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio 2019, 10, e00853-19. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, T.R.; Wu, Y. China bans colistin as a feed additive for animals. Lancet Infect. Dis. 2016, 16, 1102–1103. [Google Scholar] [CrossRef]
- Seal, B.S.; Drider, D.; Oakley, B.B.; Brüssow, H.; Bikard, D.; Rich, J.O.; Miller, S.; Devillard, E.; Kwan, J.; Bertin, G.; et al. Microbial-derived products as potential new antimicrobials. Vet. Res. 2018, 9, 66. [Google Scholar] [CrossRef] [Green Version]
- Drider, D.; Rebuffat, S. Prokaryotic Antimicrobial Peptides: From Genes to Applications; Springer: New York, NY, USA, 2011; p. 451. [Google Scholar]
- Stern, N.J.; Svetoch, E.A.; Eruslanov, B.V.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Pokhilenko, V.D.; Levchuk, V.P.; Svetoch, O.E.; Seal, B.S. Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob. Agents Chemother. 2006, 50, 3111–3116. [Google Scholar] [CrossRef] [Green Version]
- Line, J.E.; Svetoch, E.A.; Eruslanov, B.V.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Levchuk, V.P.; Svetoch, O.E.; Seal, B.S.; Siragusa, G.R.; et al. Isolation and purification of enterocin E-760 with broad antimicrobial activity against gram-positive and Gram-negative bacteria. Antimicrob. Agents Chemother. 2008, 52, 1094–1100. [Google Scholar] [CrossRef] [Green Version]
- Svetoch, E.A.; Eruslanov, B.V.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Borzenkov, V.N.; Levchuk, V.P.; Svetoch, O.E.; Kovalev, Y.N.; Stepanshin, Y.G.; et al. Diverse antimicrobial killing by Enterococcus faecium E 50-52 bacteriocin. J. Agric. Food Chem. 2008, 56, 1942–1948. [Google Scholar] [CrossRef]
- Gálvez, A.; Abriouel, H.; López, R.L.; Ben Omar, N. Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 2007, 120, 51–70. [Google Scholar] [CrossRef]
- Naghmouchi, K.; Belguesmia, Y.; Baah, J.; Teather, R.; Drider, D. Antibacterial activity of class I and IIa bacteriocins combined with polymyxin E against resistant variants of Listeria monocytogenes and Escherichia coli. Res. Microbiol. 2011, 162, 99–107. [Google Scholar] [CrossRef]
- Allen, H.K.; Trachsel, J.; Looft, T.; Casey, T.A. Finding alternatives to antibiotics: Finding alternatives to antibiotics. Ann. N. Y. Acad. Sci. 2014, 1323, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Al Atya, A.K.; Abriouel, H.; Kempf, I.; Jouy, E.; Auclair, E.; Vachée, A.; Drider, D. Effects of Colistin and Bacteriocins Combinations on the In Vitro Growth of Escherichia coli Strains from Swine Origin. Probiotics Antimicrob. Proteins 2016, 8, 183–190. [Google Scholar] [CrossRef]
- Perez, R.H.; Zendo, T.; Sonomoto, K. Novel bacteriocins from lactic acid bacteria (LAB): Various structures and applications. Microb. Cell Fact. 2014, 13 (Suppl. 1), S3. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.; Zhao, H.; Zhang, C.; Yu, J.; Lu, Z. Purification and characterization of plantaricin 163, a novel bacteriocin produced by Lactobacillus plantarum 163 isolated from traditional Chinese fermented vegetables. J. Agric. Food Chem. 2013, 61, 11676–11682. [Google Scholar] [CrossRef]
- Magnusson, J.; Schnürer, J. Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad-spectrum proteinaceous antifungal compound. Appl. Environ. Microbiol 2001, 67, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Walkenhorst, W.F.; Klein, J.W.; Vo, P.; Wimley, W.C. pH Dependence of microbe sterilization by cationic antimicrobial peptides. Antimicrob. Agents Chemother. 2013, 57, 3312–3320. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.j.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2016, 26, R14–R19. [Google Scholar] [CrossRef]
- Batdorj, B.; Dalgalarrondo, M.; Choiset, Y.; Pedroche, J.; Métro, F.; Prévost, H.; Chobert, J.M.; Haertlé, T. Purification and characterization of two bacteriocins produced by lactic acid bacteria isolated from Mongolian airag. J. Appl. Microbiol. 2006, 101, 837–848. [Google Scholar] [CrossRef]
- EUCAST. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf (accessed on 30 October 2019).
- Matuschek, E.; Åhman, J.; Webster, C.; Kahlmeter, G. Antimicrobial susceptibility testing of colistin—Evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin. Microbiol. Infect. 2018, 24, 865–870. [Google Scholar] [CrossRef] [Green Version]
- CLSI. Clinical and Laboratory Standard Institute Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Third Informational Supplement; CLSI Document VET01-S3; CLSI: Wayne, PA, USA, 2015. [Google Scholar]
- CLSI. Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Seventh Informational Supplement; CLSI Document M100-S27; CLSI: Wayne, PA, USA, 2016. [Google Scholar]
- Cavera, V.L.; Arthur, T.D.; Kashtanov, D.; Chikindas, M.L. Bacteriocins and their position in the next wave of conventional antibiotics. Int. J. Antimicrob. Agents. 2015, 46, 494–501. [Google Scholar] [CrossRef]
- Ahmad, V.; Khan, M.S.; Jamal, Q.M.S.; Alzohairy, M.A.; Al Karaawi, M.A.; Siddiqui, M.U. Antimicrobial potential of bacteriocins: In therapy, agriculture and food preservation. Int. J. Antimicrob. Agents. 2017, 49, 1–11. [Google Scholar] [CrossRef]
- Ołdak, A.; Zielińska, D. Bacteriocins from lactic acid bacteria as an alternative to antibiotics. Postepy Hig. Med. Dosw. 2017, 71, 328–338. [Google Scholar] [CrossRef]
- Mathur, H.; Field, D.; Rea, M.C.; Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocin-Antimicrobial Synergy: A Medical and Food Perspective. Front. Microbiol. 2017, 8, 1205. [Google Scholar] [CrossRef] [Green Version]
- Van Heel, A.J.; Montalban-Lopez, M.; Kuipers, O.P. Evaluating the feasibility of lantibiotics as an alternative therapy against bacterial infections in humans. Expert. Opin. Drug. Metab. Toxicol. 2011, 7, 675–680. [Google Scholar] [CrossRef] [Green Version]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Khan, S.; Tøndervik, A.; Sletta, H.; Klinkenberg, G.; Emanuel, C.; Onsøyen, E.; Myrvold, R.; Howe, R.A.; Walsh, T.R.; Hill, K.E.; et al. Overcoming drug resistance with alginate oligosaccharides able to potentiate the action of selected antibiotics. Antimicrob. Agents Chemother. 2012, 56, 5134–5141. [Google Scholar] [CrossRef] [Green Version]
- De Man, J.C.; Rogosa, M.; Sharpe, M.E. A Medium for the Cultivation of Lactobacilli. J. Appl. Bact. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Al Atya, A.K.; Belguesmia, Y.; Chataigne, G.; Ravallec, R.; Vachée, A.; Szunerits, S.; Boukherroub, R.; Drider, D. Anti-MRSA Activities of Enterocins DD28 and DD93 and Evidences on Their Role in the Inhibition of Biofilm Formation. Front. Microbiol. 2016, 31, 817. [Google Scholar] [CrossRef] [Green Version]







| Alginate Nanoparticles | Size by DLS (nm) | Zeta Potential (ζ) (mV) |
|---|---|---|
| Alg NPs (500 µg/mL) (pH 7) | 111 | −32 |
| Alg NPs (500 µg/mL) (pH 5) | 119 | −12 |
| Alg NPs + E20 fraction (pH 5) | 124 | 0 |
| pH Value | Alg NPs/E20 (500/60) µg/mL |
|---|---|
| 4 | 1–3 mm |
| 5 | 3 to 6 mm |
| 6 | 0–1 mm |
| 7 | 0–1 mm |
| Strains | Colistin (MIC µg/mL) | Fraction E20 (MIC µg/mL) | Alginate Nanoparticles (500 µg/mL) + E20 (MIC µg/mL) |
|---|---|---|---|
| E. coli 184 | 8 | 1000 | 4 |
| E. coli 289 | 8 | 2000 | 4 |
| E. coli ATCC8739 | 2 | 1000 | 2 |
| E. coli SBS36 | 2 | 1000 | 2 |
| E. coli TOP10 | 1 | 250 | 2 |
| E. coli E4A4 | 2 | 1000 | 2 |
| E. coli E5A16 | 2 | 1000 | 2 |
| Strains | Source/Origin |
|---|---|
| Lacticaseibacillus paracasei CNCM I-5369 | Traditional Algerian cheese |
| E. coli 184 * | Al Atya et al. [25] |
| E. coli 289 * | Al Atya et al. [25] |
| E. coli ATCC8739 | American Type Cell Collection, Manassas, VI (USA) |
| E. coli CIP 7624 | Collection de l’Institut Pasteur, Paris (France) |
| E. coli SBS363 | CEA, Saclay (France) |
| E. coli E4A4 | ANSES, Ploufragan (France), from healthy pig feces |
| E. coli E5A16 | ANSES, Ploufragan (France), from healthy pig feces |
| E. coli Top 10 | Invitrogen, Carlsbad, CA (USA) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belguesmia, Y.; Hazime, N.; Kempf, I.; Boukherroub, R.; Drider, D. New Bacteriocins from Lacticaseibacillus paracasei CNCM I-5369 Adsorbed on Alginate Nanoparticles Are Very Active against Escherichia coli. Int. J. Mol. Sci. 2020, 21, 8654. https://doi.org/10.3390/ijms21228654
Belguesmia Y, Hazime N, Kempf I, Boukherroub R, Drider D. New Bacteriocins from Lacticaseibacillus paracasei CNCM I-5369 Adsorbed on Alginate Nanoparticles Are Very Active against Escherichia coli. International Journal of Molecular Sciences. 2020; 21(22):8654. https://doi.org/10.3390/ijms21228654
Chicago/Turabian StyleBelguesmia, Yanath, Noura Hazime, Isabelle Kempf, Rabah Boukherroub, and Djamel Drider. 2020. "New Bacteriocins from Lacticaseibacillus paracasei CNCM I-5369 Adsorbed on Alginate Nanoparticles Are Very Active against Escherichia coli" International Journal of Molecular Sciences 21, no. 22: 8654. https://doi.org/10.3390/ijms21228654
APA StyleBelguesmia, Y., Hazime, N., Kempf, I., Boukherroub, R., & Drider, D. (2020). New Bacteriocins from Lacticaseibacillus paracasei CNCM I-5369 Adsorbed on Alginate Nanoparticles Are Very Active against Escherichia coli. International Journal of Molecular Sciences, 21(22), 8654. https://doi.org/10.3390/ijms21228654