Identification and Characterization of PLATZ Transcription Factors in Wheat
Abstract
:1. Introduction
2. Results
2.1. Identification of the PLATZ Genes in Wheat
2.2. Phylogenetic Tree and Conserved Motif Characterization of TaPLATZ Genes
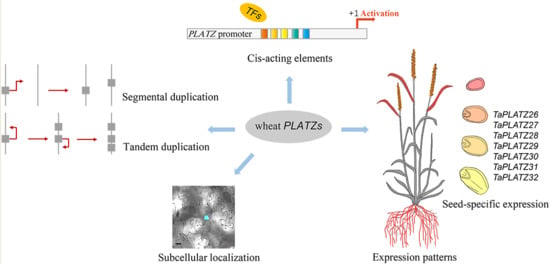
2.3. Collinearity Analysis and Gene Duplication of TaPLATZs
2.4. Variety of Cis-Acting Elements in Promoter Regions of TaPLATZs
2.5. Expression Patterns of TaPLATZs
2.6. Subcellular Localization of TaPLATZ Proteins
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. Identification of TaPLATZ Family Members in the Wheat
4.3. Characterization of TaPLATZ: Conserved Motif, PLATZ Domain and Putative Cis-Acting Elements
4.4. Phylogenetic Analysis, Collinear Relationships and Classification of PLATZ Genes in Wheat, Maize and Rice
4.5. Location of TaPLATZ Genes on the Chromosome; Identification of Duplication Genes
4.6. Expression Profiles of TaPLATZ
4.7. Subcellular Localization of TaPLATZ
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DPA | Days post-anthesis |
| HAI | Hours after imbibition |
| PLATZ | plant AT-rich protein and zinc-binding protein |
| NJ | Neighbor-Joining |
| TFs | Transcription factors |
References
- Ning, P.; Liu, C.C.; Kang, J.Q.; Lv, J.Y. Genome-wide analysis of WRKY transcription factors in wheat (Triticum aestivum L.) and differential expression under water deficit condition. PeerJ 2017, 5, e3232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.; Robatzek, S.; Somssich IEulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Kizis, D.; Lumbreras, V.; Pagès, M. Role of AP2/EREBP transcription factors in gene regulation during abiotic stress. FEBS Lett. 2001, 498, 187–189. [Google Scholar] [CrossRef] [Green Version]
- Yukio, N.; Hirofumi, F.; Takehito, I.; Yukiko, S. A novel class of plant-specific zinc-dependent DNA-binding protein that binds to A/T-rich DNA sequences. Nucleic Acids Res. 2001, 29, 4097–4105. [Google Scholar] [CrossRef] [Green Version]
- So, H.A.; Choi, S.J.; Chung, E.; Lee, J.H. Molecular characterization of stress-inducible PLATZ gene from soybean (Glycine max L.). Plant Omics 2015, 8, 479–484. [Google Scholar]
- Li, Q.; Wang, J.; Ye, J.; Zheng, X.; Xiang, X.; Li, C.; Fu, M.; Wang, Q.; Zhang, Z.; Wu, Y. The Maize Imprinted Gene Floury3 Encodes a PLATZ Protein Required for tRNA and 5S rRNA Transcription through Interaction with RNA Polymerase III. Plant Cell 2017, 29, 2661–2675. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Kim, J.; Jun, S.E.; Park, S.; Timilsina, R.; Kwon, D.S.; Kim, Y.; Park, S.-J.; Hwang, J.Y.; Nam, H.G.; et al. ORESARA15, a PLATZ transcription factor, mediates leaf growth and senescence in Arabidopsis. New Phytol. 2018, 220, 609–623. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Hou, Q.; Si, L.; Huang, X.; Luo, J.; Lu, D.; Zhu, J.; Shangguan, Y.; Miao, J.; Xie, Y.; et al. The PLATZ Transcription Factor GL6 Affects Grain Length and Number in Rice. Plant Physiol. 2019, 180, 2077–2090. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.R.; Xue, H.W. The rice PLATZ protein SHORT GRAIN6 determines grain size by regulating spikelet hull cell division. J. Integr. Plant Biol. 2019, 62, 847–864. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ji, C.; Li, Q.; Zhou, Y.; Wu, Y. Genome-wide analysis of the plant-specific PLATZ proteins in maize and identification of their general role in interaction with RNA polymerase III complex. BMC Plant Biol. 2018, 18, 221. [Google Scholar] [CrossRef] [PubMed]
- Heun, M.; SchaferPregl, R.; Klawan, D.; Castagna, R.; Accerbi, M.; Borghi, B.; Salamini, F. Site of einkorn wheat domestication identified by DNA fingerprinting. Science 1997, 278, 1312–1314. [Google Scholar] [CrossRef] [Green Version]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; Jakobsen, K.S.; Wulff, B.B.H.; Steuernagel, B.; Mayer, K.F.X.; Olsen, O.A. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, Y.; Nasuda, S. Durum wheat as a candidate for the unknown female progenitor of bread wheat: An empirical study with a highly fertile F1 hybrid with Aegilops tauschii Coss. Theor. Appl. Genet. 2004, 109, 1710–1717. [Google Scholar] [CrossRef]
- Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, 661–674. [Google Scholar] [CrossRef] [Green Version]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef] [Green Version]
- Koenig, D.; Jimenez-Gomez, J.M.; Kimura, S.; Fulop, D.; Chitwood, D.H.; Headland, L.R.; Kumar, R.; Covington, M.F.; Devisetty, U.K.; Tat, A.V.; et al. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc. Natl. Acad. Sci. USA 2013, 110, E2655–E2662. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.-X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef] [Green Version]
- Uauy, C.; Wulff, B.B.H.; Dubcovsky, J. Combining Traditional Mutagenesis with New High-Throughput Sequencing and Genome Editing to Reveal Hidden Variation in Polyploid Wheat. Annu. Rev. Genet. 2017, 51, 435–454. [Google Scholar] [CrossRef] [Green Version]
- Conte, M.G.; Gaillard, S.; Lanau, N.; Rouard, M.; Perin, C. GreenPhylDB: A database for plant comparative genomics. Nucleic Acids Res. 2007, 36, D991–D998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications may have facilitated adaptation to different environmental conditions. New Phytol. 2019, 225, 511–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurles, M. Gene duplication: The genomic trade in spare parts. PLoS Biol. 2004, 2, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 109, 217–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reidt, W.; Wohlfarth, T.; Ellerström, M.; Czihal, A.; Bäumlein, H. Gene regulation during late embryogenesis: The RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J. 2000, 21, 401–408. [Google Scholar] [CrossRef]
- Otto, S.P.; Whitton, J. Polyploid incidence and evolution. Annu. Rev. Genet. 2000, 34, 401–437. [Google Scholar] [CrossRef] [Green Version]
- Blanc, G. A Recent Polyploidy Superimposed on Older Large-Scale Duplications in the Arabidopsis Genome. Genome Res. 2003, 13, 137–144. [Google Scholar] [CrossRef] [Green Version]
- Panchy, N.; Lehti-Shiu, M.D.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [Green Version]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [Green Version]
- Feldman, M.; Levy, A.A. Genome Evolution Due to Allopolyploidization in Wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, N.N.; Deng, X.; Liu, D.M.; Li, M.F.; Cui, D.D.; Hu, Y.K.; Yan, Y.M. Genome-wide analysis of wheat DNA-binding with one finger (Dof) transcription factor genes: Evolutionary characteristics and diverse abiotic stress responses. BMC Genom. 2020, 21, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Rahman, S.; Islam, S.; Yu, Z.; She, M.; Nevo, E.; Ma, W. Current Progress in Understanding and Recovering the Wheat Genes Lost in Evolution and Domestication. Int. J. Mol. Sci. 2020, 21, 5836. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; Pires, J.C. Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009, 17, 699–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; DePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.L.; Wendel, J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005, 8, 135–141. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar]
- Lynch, M.; Force, A. The probability of duplicate gene preservation by subfunctionalization. Genetics 2000, 154, 459–473. [Google Scholar]
- Kiełbowicz-Matuk, A. Involvement of plant C2H2-type zinc finger transcription factors in stress responses. Plant Sci. Int. J. Exp. Plant Biol. 2012, 185–186, 78–85. [Google Scholar] [CrossRef]
- González-Morales, S.I.; Chávez-Montes, R.A.; Hayano-Kanashiro, C.; Alejo-Jacuinde, G.; Rico-Cambron, T.Y.; de Folter, S.; Herrera-Estrella, L. Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2016, 113, E5232. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Yang, R.; Huo, Y.; Liu, S.; Yang, G.; Huang, J.; Zheng, C.; Wu, C. Expression of cotton PLATZ1 in transgenic Arabidopsis reduces sensitivity to osmotic and salt stress for germination and seedling establishment associated with modification of the abscisic acid, gibberellin, and ethylene signalling pathways. BMC Plant Biol. 2018, 18, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinerson, C.I.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Rushton, P.J. The evolution of WRKY transcription factors. BMC Plant Biol. 2015, 15, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Magali, L. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 1, 325–327. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Yue, H.; Feng, K.; Deng, P.; Song, W.; Nie, X. Genome-wide identification, phylogeny and expressional profiles of mitogen activated protein kinase kinase kinase (MAPKKK) gene family in bread wheat (Triticum aestivum L.). BMC Genom. 2016, 17, 668. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Ma, R.; Xu, D.; Bi, H.; Xia, Z.; Peng, H. Genome-Wide Identification and Analysis of the AP2 Transcription Factor Gene Family in Wheat (Triticum aestivum L.). Front. Plant Sci. 2019, 10, 1286. [Google Scholar] [CrossRef] [Green Version]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Bi, C.; Xu, Y.; Ye, Q.; Yin, T.; Ye, N. Genome-wide identification and characterization of WRKY gene family inSalix suchowensis. PeerJ 2016, 4, e2437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarosová, J.; Kundu, J.K. Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol. 2010, 10, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, X.-Y.; Wang, J.-R.; Ouellet, T.; Rocheleau, H.; Wei, Y.-M.; Pu, Z.-E.; Jiang, Q.-T.; Lan, X.-J.; Zheng, Y.-L. Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol. Biol. 2010, 74, 307–311. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yu, C.-S.; Chen, Y.-C.; Lu, C.-H.; Hwang, J.-K. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Y.; Cheng, M.; Li, M.; Guo, X.; Wu, Y.; Wang, J. Identification and Characterization of PLATZ Transcription Factors in Wheat. Int. J. Mol. Sci. 2020, 21, 8934. https://doi.org/10.3390/ijms21238934
Fu Y, Cheng M, Li M, Guo X, Wu Y, Wang J. Identification and Characterization of PLATZ Transcription Factors in Wheat. International Journal of Molecular Sciences. 2020; 21(23):8934. https://doi.org/10.3390/ijms21238934
Chicago/Turabian StyleFu, Yuxin, Mengping Cheng, Maolian Li, Xiaojiang Guo, Yongrui Wu, and Jirui Wang. 2020. "Identification and Characterization of PLATZ Transcription Factors in Wheat" International Journal of Molecular Sciences 21, no. 23: 8934. https://doi.org/10.3390/ijms21238934
APA StyleFu, Y., Cheng, M., Li, M., Guo, X., Wu, Y., & Wang, J. (2020). Identification and Characterization of PLATZ Transcription Factors in Wheat. International Journal of Molecular Sciences, 21(23), 8934. https://doi.org/10.3390/ijms21238934