The Evolutionary Significance of RNAi in the Fungal Kingdom
Abstract
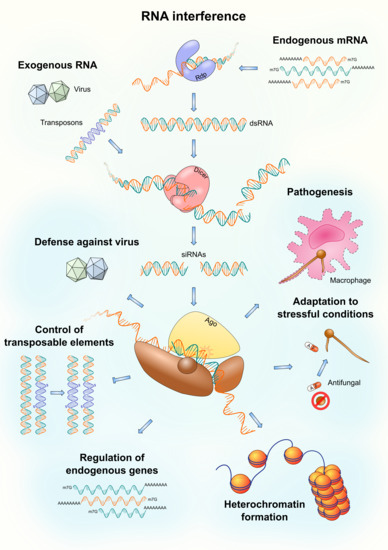
:1. Introduction
2. Defense against Viruses
3. Control of Transposable Elements
4. Regulation of Endogenous Genes
5. Heterochromatin Formation
6. Adaptation to Stressful Conditions
7. Pathogenesis
8. Loss of RNAi
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| RNAi | RNA interference |
| dsRNA | double-stranded RNA |
| siRNAs | small interfering RNAs |
| sRNAs | small RNAs |
| RISC | RNA-induced silencing complex |
| RITS | RNA-induced transcriptional silencing complex |
| ssRNA | single-stranded RNA |
| RdRP/Rdp | RNA-dependent RNA polymerase |
| PTGS | Post-transcriptional gene silencing |
| MSUD | Meiotic silencing of unpaired DNA |
| masiRNAs | MSUD-associated siRNAs |
| vsRNAs | virus-derived small interfering RNAs |
| TE | transposable elements |
| SCANR | spliceosome-coupled and nuclear RNAi |
| TUTases | terminal-uridylyl transferases |
| miRNAs | micro RNAs |
| milRNAs | microRNA-like RNAs |
| NCRIP | noncanonical RNAi pathway |
| ex-siRNAs | exonic siRNAs |
| rdRNAs | RdRP-dependent dicer-independent sRNAs |
| disiRNAs | Dicer-independent small interfering RNAs |
| DLDM | disiRNA loci DNA methylation |
| tRFs | tRNA fragments |
| circRNAs | circular RNAs |
| CLRC | cryptic loci regulator complex |
| H3K9me | methylation of the histone 3 lysin 9 |
| 5-FOA | 5-fluoroorotic acid |
References
- Chang, S.S.; Zhang, Z.; Liu, Y. RNA interference pathways in fungi: Mechanisms and functions. Annu. Rev. Microbiol. 2012, 66, 305–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerutti, H.; Casas-Mollano, J.A. On the origin and functions of RNA-mediated silencing: From protists to man. Curr. Genet. 2006, 50, 91–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matveyev, A.V.; Alves, J.M.P.; Serrano, M.G.; Lee, V.; Lara, A.M.; Barton, W.A.; Costa-Martins, A.G.; Beverley, S.M.; Camargo, E.P.; Teixeira, M.M.G.; et al. The Evolutionary Loss of RNAi Key Determinants in Kinetoplastids as a Multiple Sporadic Phenomenon. J. Mol. Evol. 2017, 84, 104–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ketting, R.F. The Many Faces of RNAi. Dev. Cell 2011, 20, 148–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kataoka, K.; Mochizuki, K. Programmed DNA elimination in tetrahymena: A small RNA-mediated genome surveillance mechanism. Adv. Exp. Med. Biol. 2011, 722, 156–173. [Google Scholar] [CrossRef] [Green Version]
- Nicolás, F.E.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Loss and Retention of RNA Interference in Fungi and Parasites. PLoS Pathog. 2013, 9, e1003089. [Google Scholar] [CrossRef] [Green Version]
- Sarkies, P.; Selkirk, M.E.; Jones, J.T.; Blok, V.; Boothby, T.; Goldstein, B.; Hanelt, B.; Ardila-Garcia, A.; Fast, N.M.; Schiffer, P.M.; et al. Ancient and Novel Small RNA Pathways Compensate for the Loss of piRNAs in Multiple Independent Nematode Lineages. PLoS Biol. 2015, 13, e1002061. [Google Scholar] [CrossRef] [Green Version]
- Tomoyasu, Y.; Miller, S.C.; Tomita, S.; Schoppmeier, M.; Grossmann, D.; Bucher, G. Exploring systemic RNA interference in insects: A genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008, 9, R10. [Google Scholar] [CrossRef] [Green Version]
- Dowling, D.; Pauli, T.; Donath, A.; Meusemann, K.; Podsiadlowski, L.; Petersen, M.; Peters, R.S.; Mayer, C.; Liu, S.; Zhou, X.; et al. Phylogenetic origin and diversification of RNAi pathway genes in insects. Genome Biol. Evol. 2016, 8, 3784–3793. [Google Scholar] [CrossRef] [Green Version]
- Hedil, M.; Kormelink, R. Viral RNA silencing suppression: The enigma of bunyavirus NSs proteins. Viruses 2016, 8, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muhammad, T.; Zhang, F.; Zhang, Y.; Liang, Y. RNA Interference: A Natural Immune System of Plants to Counteract Biotic Stressors. Cells 2019, 8, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Danielsen, J.M.R.; Yang, Y.G.; Qi, Y. A role for small RNAs in DNA double-strand break repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svoboda, P. Renaissance of mammalian endogenous RNAi. FEBS Lett. 2014, 588, 2550–2556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, N.; Macino, G. Quelling: Transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 1992, 6, 3343–3353. [Google Scholar] [CrossRef]
- Cogoni, C.; Macino, G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 1999, 399, 166–169. [Google Scholar] [CrossRef]
- Nakayashiki, H. RNA silencing in fungi: Mechanisms and applications. FEBS Lett. 2005, 579, 5950–5957. [Google Scholar] [CrossRef] [Green Version]
- Son, H.; Min, K.; Lee, J.; Raju, N.B.; Lee, Y.W. Meiotic silencing in the homothallic fungus Gibberella zeae. Fungal Biol. 2011, 12, 1290–1302. [Google Scholar] [CrossRef]
- Wang, Y.; Smith, K.M.; Taylor, J.W.; Freitag, M.; Stajich, J.E. Endogenous small RNA mediates meiotic silencing of a novel DNA transposon. G3 Genes Genomes Genet. 2015, 5, 1949–1960. [Google Scholar] [CrossRef] [Green Version]
- Decker, L.M.; Boone, E.C.; Xiao, H.; Shanker, B.S.; Boone, S.F.; Kingston, S.L.; Lee, S.A.; Hammond, T.M.; Shiu, P.K.T. Complex formation of RNA silencing proteins in the perinuclear region of Neurospora crassa. Genetics 2015, 199, 1017–1021. [Google Scholar] [CrossRef] [Green Version]
- Hirano, Y.; Asakawa, H.; Sakuno, T.; Haraguchi, T.; Hiraoka, Y. Nuclear Envelope Proteins Modulating the Heterochromatin Formation and Functions in Fission Yeast. Cells 2020, 9, 1908. [Google Scholar] [CrossRef] [PubMed]
- Billmyre, R.B.; Calo, S.; Feretzaki, M.; Wang, X.; Heitman, J. RNAi function, diversity, and loss in the fungal kingdom. Chromosom. Res. 2013, 21, 561–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres-Martínez, S.; Ruiz-Vázquez, R.M. The RNAi Universe in Fungi: A Varied Landscape of Small RNAs and Biological Functions. Annu. Rev. Microbiol. 2017, 71, 371–391. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Choi, G.H.; Nuss, D.L. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. USA 2009, 106, 17927–17932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Nuss, D.L. A host dicer is required for defective viral RNA production and recombinant virus vector RNA instability for a positive sense RNA virus. Proc. Natl. Acad. Sci. USA 2008, 105, 16749–16754. [Google Scholar] [CrossRef] [Green Version]
- Hammond, T.M.; Andrewski, M.D.; Roossinck, M.J.; Keller, N.P. Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryot. Cell 2008, 7, 350–357. [Google Scholar] [CrossRef] [Green Version]
- Nolan, T.; Braccini, L.; Azzalin, G.; De Toni, A.; Macino, G.; Cogoni, C. The post-transcriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa. Nucleic Acids Res. 2005, 33, 1564–1573. [Google Scholar] [CrossRef]
- Shiu, P.K.T.; Raju, N.B.; Zickler, D.; Metzenberg, R.L. Meiotic silencing by unpaired DNA. Cell 2001, 107, 905–916. [Google Scholar] [CrossRef] [Green Version]
- Yamanaka, S.; Mehta, S.; Reyes-Turcu, F.E.; Zhuang, F.; Fuchs, R.T.; Rong, Y.; Robb, G.B.; Grewal, S.I.S. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature 2013, 493, 557–560. [Google Scholar] [CrossRef] [Green Version]
- Nunes, C.C.; Gowda, M.; Sailsbery, J.; Xue, M.; Chen, F.; Brown, D.E.; Oh, Y.Y.; Mitchell, T.K.; Dean, R.A. Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae. BMC Genom. 2011, 12, 288. [Google Scholar] [CrossRef] [Green Version]
- Kadotani, N.; Nakayashiki, H.; Tosa, Y.; Mayama, S. RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2003, 16, 769–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murata, T.; Kadotani, N.; Yamaguchi, M.; Tosa, Y.; Mayama, S.; Nakayashiki, H. siRNA-dependent and -independent post-transcriptional cosuppression of the LTR-retrotransposon MAGGY in the phytopathogenic fungus Magnaporthe oryzae. Nucleic Acids Res. 2007, 35, 5987–5994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayashiki, H.; Ikeda, K.; Hashimoto, Y.; Tosa, Y.; Mayama, S. Methylation is not the main force repressing the retrotransposon MAGGY in Magnaporthe grisea. Nucleic Acids Res. 2001, 29, 1278–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro-Mendoza, M.I.; Pérez-Arques, C.; Panchal, S.; Nicolás, F.E.; Mondo, S.J.; Ganguly, P.; Pangilinan, J.; Grigoriev, I.V.; Heitman, J.; Sanyal, K.; et al. Early Diverging Fungus Mucor circinelloides Lacks Centromeric Histone CENP-A and Displays a Mosaic of Point and Regional Centromeres. Curr. Biol. 2019, 29, 3791–3802.e6. [Google Scholar] [CrossRef] [Green Version]
- Janbon, G.; Maeng, S.; Yang, D.H.; Ko, Y.J.; Jung, K.W.; Moyrand, F.; Floyd, A.; Heitman, J.; Bahn, Y.S. Characterizing the role of RNA silencing components in Cryptococcus neoformans. Fungal Genet. Biol. 2010, 47, 1070. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Hsueh, Y.P.; Li, W.; Floyd, A.; Skalsky, R.; Heitman, J. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 2010, 24, 2566–2582. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Darwiche, S.; Heitman, J. Sex-induced silencing operates during opposite-sex and unisexual reproduction in Cryptococcus neoformans. Genetics 2013, 193, 1163–1174. [Google Scholar] [CrossRef] [Green Version]
- Dumesic, P.A.; Natarajan, P.; Chen, C.; Drinnenberg, I.A.; Schiller, B.J.; Thompson, J.; Moresco, J.J.; Yates, J.R.; Bartel, D.P.; Madhani, H.D. Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell 2013, 152, 957–968. [Google Scholar] [CrossRef] [Green Version]
- Burke, J.E.; Longhurst, A.D.; Natarajan, P.; Rao, B.; Liu, J.; Sales-Lee, J.; Mortensen, Y.; Moresco, J.J.; Diedrich, J.K.; Yates, J.R.; et al. A non-dicer RNase III and four other novel factors required for RNAi-Mediated transposon suppression in the human pathogenic yeast cryptococcus neoformans. G3 Genes Genomes Genet. 2019, 9, 2235–2244. [Google Scholar] [CrossRef] [Green Version]
- Sperschneider, J.; Jones, A.W.; Nasim, J.; Xu, B.; Jacques, S.; Upadhyaya, N.M.; Mago, R.; Figueroa, M.; Singh, K.B.; Stone, E.A.; et al. The stem rust fungus Puccinia graminis f. sp. tritici induces centromeric small RNAs during late infection that direct genome-wide DNA methylation. bioRxiv 2020, 469338. [Google Scholar] [CrossRef] [Green Version]
- Nicolás, F.E.; Vila, A.; Moxon, S.; Cascales, M.D.; Torres-Martínez, S.; Ruiz-Vázquez, R.M.; Garre, V. The RNAi machinery controls distinct responses to environmental signals in the basal fungus Mucor circinelloides. BMC Genom. 2015, 16, 237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calo, S.; Nicolás, F.E.; Lee, S.C.; Vila, A.; Cervantes, M.; Torres-Martinez, S.; Ruiz-Vazquez, R.M.; Cardenas, M.E.; Heitman, J. A non-canonical RNA degradation pathway suppresses RNAi-dependent epimutations in the human fungal pathogen Mucor circinelloides. PLoS Genet. 2017, 13, e1006686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trieu, T.A.; Calo, S.; Nicolás, F.E.; Vila, A.; Moxon, S.; Dalmay, T.; Torres-Martínez, S.; Garre, V.; Ruiz-Vázquez, R.M. A Non-canonical RNA Silencing Pathway Promotes mRNA Degradation in Basal Fungi. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carreras-Villaseñor, N.; Esquivel-Naranjo, E.U.; Villalobos-Escobedo, J.M.; Abreu-Goodger, C.; Herrera-Estrella, A. The RNAi machinery regulates growth and development in the filamentous fungus Trichoderma atroviride. Mol. Microbiol. 2013, 89, 96–112. [Google Scholar] [CrossRef] [PubMed]
- Son, H.; Park, A.R.; Lim, J.Y.; Shin, C.; Lee, Y.W. Genome-wide exonic small interference RNA-mediated gene silencing regulates sexual reproduction in the homothallic fungus Fusarium graminearum. PLoS Genet. 2017, 13, e1006595. [Google Scholar] [CrossRef]
- Zeng, W.; Wang, J.; Wang, Y.; Lin, J.; Fu, Y.; Xie, J.; Jiang, D.; Chen, T.; Liu, H.; Cheng, J. Dicer-like proteins regulate sexual development via the biogenesis of perithecium-specific microRNAs in a plant pathogenic fungus Fusarium graminearum. Front. Microbiol. 2018, 9, 818. [Google Scholar] [CrossRef] [Green Version]
- Lau, A.Y.T.; Cheng, X.; Cheng, C.K.; Nong, W.; Cheung, M.K.; Chan, R.H.F.; Hui, J.H.L.; Kwan, H.S. Discovery of microRNA-like RNAs during early fruiting body development in the model mushroom Coprinopsis cinerea. PLoS ONE 2018, 13, e0198234. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.C.; Li, L.; Gu, W.; Xue, Z.; Crosthwaite, S.K.; Pertsemlidis, A.; Lewis, Z.A.; Freitag, M.; Selker, E.U.; Mello, C.C.; et al. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol. Cell 2010, 38, 803–814. [Google Scholar] [CrossRef] [Green Version]
- Dang, Y.; Cheng, J.; Sun, X.; Zhou, Z.; Liu, Y. Antisense transcription licenses nascent transcripts to mediate transcriptional gene silencing. Genes Dev. 2016, 30, 2417–2432. [Google Scholar] [CrossRef] [Green Version]
- Dang, Y.; Li, L.; Guo, W.; Xue, Z.; Liu, Y. Convergent Transcription Induces Dynamic DNA Methylation at disiRNA Loci. PLoS Genet. 2013, 9, e1003761. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Wang, Z.; Xing, J.; Yang, Q.; Chen, X.L. Genome-wide Identification and characterization of circular RNAs in the rice blast fungus Magnaporthe oryzae. Sci. Rep. 2018, 8, 6757. [Google Scholar] [CrossRef] [PubMed]
- Åsman, A.K.M.; Vetukuri, R.R.; Jahan, S.N.; Fogelqvist, J.; Corcoran, P.; Avrova, A.O.; Whisson, S.C.; Dixelius, C. Fragmentation of tRNA in Phytophthora infestans asexual life cycle stages and during host plant infection. BMC Microbiol. 2014, 14, 308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martienssen, R.; Moazed, D. RNAi and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 2015, 7, a019323. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Wang, X.; Moazed, D. Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation. Nature 2018, 558, 615–619. [Google Scholar] [CrossRef]
- Calo, S.; Shertz-Wall, C.; Lee, S.C.; Bastidas, R.J.; Nicolás, F.E.; Granek, J.A.; Mieczkowski, P.; Torres-Martínez, S.; Ruiz-Vázquez, R.M.; Cardenas, M.E.; et al. Antifungal drug resistance evoked via RNAi-dependent epimutations. Nature 2014, 513. [Google Scholar] [CrossRef] [Green Version]
- Chang, Z.; Billmyre, R.B.; Lee, S.C.; Heitman, J. Broad antifungal resistance mediated by RNAi-dependent epimutation in the basal human fungal pathogen Mucor circinelloides. PLoS Genet. 2019, 15, e1007957. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; An, B.; Hou, X.; Guo, Y.; Luo, H.; He, C. Dicer-like proteins regulate the growth, conidiation, and pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis. Front. Microbiol. 2018, 8, 2621. [Google Scholar] [CrossRef] [Green Version]
- Raman, V.; Simon, S.A.; Demirci, F.; Nakano, M.; Meyers, B.C.; Donofrio, N.M. Small RNA functions are required for growth and development of magnaporthe oryzae. Mol. Plant-Microbe Interact. 2017, 30, 517–530. [Google Scholar] [CrossRef] [Green Version]
- Mochama, P.; Jadhav, P.; Neupane, A.; Marzano, S.Y.L. Mycoviruses as triggers and targets of RNA silencing in white mold fungus Sclerotinia sclerotiorum. Viruses 2018, 10, 214. [Google Scholar] [CrossRef] [Green Version]
- Feng, H.; Xu, M.; Liu, Y.; Gao, X.; Yin, Z.; Voegele, R.T.; Huang, L. The distinct roles of Argonaute protein 2 in the growth, stress responses and pathogenicity of the apple tree canker pathogen. For. Pathol. 2017, 47, e12354. [Google Scholar] [CrossRef]
- Jo, S.M.; Ayukawa, Y.; Yun, S.H.; Komatsu, K.; Arie, T. A putative RNA silencing component protein FoQde-2 is involved in virulence of the tomato wilt fungus Fusarium oxysporum f. sp. lycopersici. J. Gen. Plant Pathol. 2018, 84, 395–398. [Google Scholar] [CrossRef]
- Yin, C.; Zhu, H.; Jiang, Y.; Shan, Y.; Gong, L. Silencing Dicer-Like Genes Reduces Virulence and sRNA Generation in Penicillium italicum, the Cause of Citrus Blue Mold. Cells 2020, 9, 363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaffar, F.Y.; Imani, J.; Karlovsky, P.; Koch, A.; Kogel, K.-H. Different Components of the RNA Interference Machinery Are Required for Conidiation, Ascosporogenesis, Virulence, Deoxynivalenol Production, and Fungal Inhibition by Exogenous Double-Stranded RNA in the Head Blight Pathogen Fusarium graminearum. Front. Microbiol. 2019, 10, 1662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Arques, C.; Navarro-Mendoza, M.I.; Murcia, L.; Navarro, E.; Garre, V.; Nicolás, F.E. A non-canonical RNAi pathway controls virulence and genome stability in Mucorales. PLoS Genet. 2020, 16, e1008611. [Google Scholar] [CrossRef]
- Qiao, Y.; Liu, L.; Xiong, Q.; Flores, C.; Wong, J.; Shi, J.; Wang, X.; Liu, X.; Xiang, Q.; Jiang, S.; et al. Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 2013, 45, 330–333. [Google Scholar] [CrossRef] [Green Version]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Sun, Y.; Song, N.; Zhao, M.; Liu, R.; Feng, H.; Wang, X.; Kang, Z. Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 2017, 215, 338–350. [Google Scholar] [CrossRef] [Green Version]
- Kusch, S.; Frantzeskakis, L.; Thieron, H.; Panstruga, R. Small RNAs from cereal powdery mildew pathogens may target host plant genes. Fungal Biol. 2018, 122, 1050–1063. [Google Scholar] [CrossRef]
- Dunker, F.; Trutzenberg, A.; Rothenpieler, J.S.; Kuhn, S.; Pröls, R.; Schreiber, T.; Tissier, A.; Kemen, A.; Kemen, E.; Hückelhoven, R.; et al. Oomycete small RNAs bind to the plant RNA-induced silencing complex for virulence. Elife 2020, 9, e56096. [Google Scholar] [CrossRef]
- Segers, G.C.; Zhang, X.; Deng, F.; Sun, Q.; Nuss, D.L. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. USA 2007, 104, 12902–12906. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, S.; Lee, H.-C.; Maiti, M.; He, Q.; Cheng, P.; Liu, Q.; Liu, Y. A Double-Stranded-RNA Response Program Important for RNA Interference Efficiency. Mol. Cell. Biol. 2007, 27, 3995–4005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segers, G.C.; Van Wezel, R.; Zhang, X.; Hong, Y.; Nuss, D.L. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell 2006, 5, 896–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotta-Loizou, I.; Coutts, R.H.A. Mycoviruses in Aspergilli: A comprehensive review. Front. Microbiol. 2017, 8, 1699. [Google Scholar] [CrossRef] [Green Version]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.J.; O’Neill, R.J. Transposable elements: Genome innovation, chromosome diversity, and centromere conflict. Chromosom. Res. 2018, 26, 5–23. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, C.D.; Springer, N.M. Transposable element influences on gene expression in plants. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Diehl, A.G.; Ouyang, N.; Boyle, A.P. Transposable elements contribute to cell and species-specific chromatin looping and gene regulation in mammalian genomes. Nat. Commun. 2020, 11, 1796. [Google Scholar] [CrossRef] [Green Version]
- Friedli, M.; Trono, D. The Developmental Control of Transposable Elements and the Evolution of Higher Species. Annu. Rev. Cell Dev. Biol. 2015, 31, 429–451. [Google Scholar] [CrossRef]
- Castanera, R.; López-Varas, L.; Borgognone, A.; LaButti, K.; Lapidus, A.; Schmutz, J.; Grimwood, J.; Pérez, G.; Pisabarro, A.G.; Grigoriev, I.V.; et al. Transposable Elements versus the Fungal Genome: Impact on Whole-Genome Architecture and Transcriptional Profiles. PLoS Genet. 2016, 12, e1006108. [Google Scholar] [CrossRef] [Green Version]
- Bourc’his, D.; Bestor, T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 2004, 431, 96–99. [Google Scholar] [CrossRef]
- Bulut-Karslioglu, A.; DeLaRosa-Velázquez, I.A.; Ramirez, F.; Barenboim, M.; Onishi-Seebacher, M.; Arand, J.; Galán, C.; Winter, G.E.; Engist, B.; Gerle, B.; et al. Suv39h-Dependent H3K9me3 Marks Intact Retrotransposons and Silences LINE Elements in Mouse Embryonic Stem Cells. Mol. Cell 2014, 55, 277–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quenneville, S.; Turelli, P.; Bojkowska, K.; Raclot, C.; Offner, S.; Kapopoulou, A.; Trono, D. The KRAB-ZFP/KAP1 System Contributes to the Early Embryonic Establishment of Site-Specific DNA Methylation Patterns Maintained during Development. Cell Rep. 2012, 2, 766–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourc’his, D.; Voinnet, O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science 2010, 330, 617–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aravin, A.A.; Naumova, N.M.; Tulin, A.V.; Vagin, V.V.; Rozovsky, Y.M.; Gvozdev, V.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001, 11, 1017–1027. [Google Scholar] [CrossRef] [Green Version]
- Sijen, T.; Plasterk, R.H.A. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 2003, 426, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Ketting, R.F.; Haverkamp, T.H.A.; Van Luenen, H.G.A.M.; Plasterk, R.H.A. mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of werner syndrome helicase and RNaseD. Cell 1999, 99, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Tabara, H.; Sarkissian, M.; Kelly, W.G.; Fleenor, J.; Grishok, A.; Timmons, L.; Fire, A.; Mello, C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 1999, 99, 123–132. [Google Scholar] [CrossRef] [Green Version]
- Wu-Scharf, D.; Jeong, B.R.; Zhang, C.; Cerutti, H. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-Box RNA helicase. Science 2000, 290, 1159–1162. [Google Scholar] [CrossRef] [Green Version]
- Soifer, H.S.; Zaragoza, A.; Peyvan, M.; Behlke, M.A.; Rossi, J.J. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res. 2005, 33, 846–856. [Google Scholar] [CrossRef] [Green Version]
- Hammond, T.M.; Bok, J.W.; Andrewski, M.D.; Reyes-Domínguez, Y.; Scazzocchio, C.; Keller, N.P. RNA silencing gene truncation in the filamentous fungus Aspergillus nidulans. Eukaryot. Cell 2008, 7, 339–349. [Google Scholar] [CrossRef] [Green Version]
- Wagner, L.; De Hoog, S.; Alastruey-Izquierdo, A.; Voigt, K.; Kurzai, O.; Walther, G. A revised species concept for opportunistic Mucor species reveals species-specific antifungal susceptibility profiles. Antimicrob. Agents Chemother. 2019, 63, e00653-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolas, F.E.; Moxon, S.; de Haro, J.P.; Calo, S.; Grigoriev, I.V.; Torres-MartÍnez, S.; Moulton, V.; Ruiz-Vázquez, R.M.; Dalmay, T. Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. 2010, 38. [Google Scholar] [CrossRef] [PubMed]
- Loftus, B.J.; Fung, E.; Roncaglia, P.; Rowley, D.; Amedeo, P.; Bruno, D.; Vamathevan, J.; Miranda, M.; Anderson, I.J.; Fraser, J.A.; et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 2005, 307, 1321–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warkocki, Z.; Krawczyk, P.S.; Adamska, D.; Bijata, K.; Garcia-Perez, J.L.; Dziembowski, A. Uridylation by TUT4/7 Restricts Retrotransposition of Human LINE-1s. Cell 2018, 174, 1537–1548.e29. [Google Scholar] [CrossRef] [Green Version]
- Jiang, N.; Yang, Y.; Janbon, G.; Pan, J.; Zhu, X. Identification and Functional Demonstration of miRNAs in the Fungus Cryptococcus neoformans. PLoS ONE 2012, 7, e52734. [Google Scholar] [CrossRef] [Green Version]
- Yadav, V.; Sun, S.; Billmyre, R.B.; Thimmappa, B.C.; Shea, T.; Lintner, R.; Bakkeren, G.; Cuomo, C.A.; Heitman, J.; Sanyal, K. RNAi is a critical determinant of centromere evolution in closely related fungi. Proc. Natl. Acad. Sci. USA 2018, 115, 3108–3113. [Google Scholar] [CrossRef] [Green Version]
- Fahlgren, N.; Bollmann, S.R.; Kasschau, K.D.; Cuperus, J.T.; Press, C.M.; Sullivan, C.M.; Chapman, E.J.; Hoyer, J.S.; Gilbert, K.B.; Grünwald, N.J.; et al. Phytophthora Have Distinct Endogenous Small RNA Populations That Include Short Interfering and microRNAs. PLoS ONE 2013, 8, e77181. [Google Scholar] [CrossRef]
- Vetukuri, R.R.; Åsman, A.K.M.; Tellgren-Roth, C.; Jahan, S.N.; Reimegård, J.; Fogelqvist, J.; Savenkov, E.; Söderbom, F.; Avrova, A.O.; Whisson, S.C.; et al. Evidence for Small RNAs Homologous to Effector-Encoding Genes and Transposable Elements in the Oomycete Phytophthora infestans. PLoS ONE 2012, 7, e51399. [Google Scholar] [CrossRef] [Green Version]
- Grandaubert, J.; Lowe, R.G.T.; Soyer, J.L.; Schoch, C.L.; Van De Wouw, A.P.; Fudal, I.; Robbertse, B.; Lapalu, N.; Links, M.G.; Ollivier, B.; et al. Transposable element-assisted evolution and adaptation to host plant within the Leptosphaeria maculans-Leptosphaeria biglobosa species complex of fungal pathogens. BMC Genomics 2014, 15, 891. [Google Scholar] [CrossRef] [Green Version]
- Lax, C.; Pérez-arques, C.; Navarro-mendoza, M.I.; Cánovas-márquez, J.T.; Tahiri, G.; Pérez-ruiz, J.A.; Osorio-concepción, M.; Murcia-flores, L.; Navarro, E.; Garre, V.; et al. Genes, pathways, and mechanisms involved in the virulence of mucorales. Genes 2020, 11, 317. [Google Scholar] [CrossRef] [Green Version]
- Nicolás, F.E.; Murcia, L.; Navarro, E.; Navarro-Mendoza, M.I.; Pérez-Arques, C.; Garre, V. Mucorales species and macrophages. J. Fungi 2020, 6, 94. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, M.; Vila, A.; Nicolas, F.E.; Moxon, S.; de Haro, J.P.; Dalmay, T.; Torres-Martinez, S.; Ruiz-Vazquez, R.M. A single argonaute gene participates in exogenous and endogenous RNAi and controls cellular functions in the basal fungus Mucor circinelloides. PLoS ONE 2013, 8, e69283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Haro, J.P.; Calo, S.; Cervantes, M.; Nicolás, F.E.; Torres-Martinez, S.; Ruiz-Vázquez, R.M. A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in mucor circinelloides. Eukaryot. Cell 2009, 8, 1486–1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolas, F.E.; de Haro, J.P.; Torres-Martinez, S.; Ruiz-Vazquez, R.M. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 2007, 44, 504–516. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Yang, Q.; Li, L.; Xue, Z.; Ye, Q.; Zhang, L.; Li, S.; Liu, Y. Transcription of the Major Neurospora crassa microRNA-Like Small RNAs Relies on RNA Polymerase III. PLoS Genet. 2013, 9, e1003227. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Gao, Q.; Huang, M.; Liu, Y.; Liu, Z.; Liu, X.; Ma, Z. Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum. Sci. Rep. 2015, 5, 12500. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef] [Green Version]
- Sobala, A.; Hutvagner, G. Small RNAs derived from the 5′ end of tRNAs can inhibit protein translation in human cells. RNA Biol. 2013, 10, 553–563. [Google Scholar] [CrossRef] [Green Version]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. TRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Ptashkin, R.N.; Chen, Y.; Cheng, Z.; Liu, G.; Phan, T.; Deng, X.; Zhou, J.; Lee, I.; Lee, Y.S.; et al. Respiratory Syncytial Virus Utilizes a tRNA Fragment to Suppress Antiviral Responses Through a Novel Targeting Mechanism. Mol. Ther. 2015, 23, 1622–1629. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Ali, M.; Zhou, Q. Establishment and evolution of heterochromatin. Ann. N. Y. Acad. Sci. 2020, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allshire, R.C.; Madhani, H.D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 2018, 19, 229–244. [Google Scholar] [CrossRef]
- Taglini, F.; Chapman, E.; van Nues, R.; Theron, E.; Bayne, E.H. Mkt1 is required for RNAi-mediated silencing and establishment of heterochromatin in fission yeast. Nucleic Acids Res. 2020, 48, 1239–1253. [Google Scholar] [CrossRef] [Green Version]
- Volpe, T.A.; Kidner, C.; Hall, I.M.; Teng, G.; Grewal, S.I.; Martienssen, R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2002, 297, 1833–1837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verdel, A.; Jia, S.; Gerber, S.; Sugiyama, T.; Gygi, S.; Grewal, S.I.; Moazed, D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 2004, 303, 672–676. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjee, S.; Roche, B.; Martienssen, R.A. RNA-induced initiation of transcriptional silencing (RITS) complex structure and function. RNA Biol. 2019, 16, 1133–1146. [Google Scholar] [CrossRef] [PubMed]
- Jih, G.; Iglesias, N.; Currie, M.A.; Bhanu, N.V.; Paulo, J.A.; Gygi, S.P.; Garcia, B.A.; Moazed, D. Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription. Nature 2017, 547, 463–467. [Google Scholar] [CrossRef] [Green Version]
- Duempelmann, L.; Skribbe, M.; Bühler, M. Small RNAs in the Transgenerational Inheritance of Epigenetic Information. Trends Genet. 2020, 36, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Thon, G.; Hansen, K.R.; Altes, S.P.; Sidhu, D.; Singh, G.; Verhein-Hansen, J.; Bonaduce, M.J.; Klar, A.J.S. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics 2005, 171, 1583–1595. [Google Scholar] [CrossRef] [Green Version]
- Hall, I.M.; Shankaranarayana, G.D.; Noma, K.; Ayoub, N.; Cohen, A.; Grewal, S.I. Establishment and maintenance of a heterochromatin domain. Science 2002, 297, 2232–2237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, D.B.; Nakayama, J.I. RNA and epigenetic silencing: Insight from fission yeast. Dev. Growth Differ. 2012, 54, 129–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadaie, M.; Kawaguchi, R.; Ohtani, Y.; Arisaka, F.; Tanaka, K.; Shirahige, K.; Nakayama, J. Balance between Distinct HP1 Family Proteins Controls Heterochromatin Assembly in Fission Yeast. Mol. Cell. Biol. 2008, 28, 6973–6988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duempelmann, L.; Mohn, F.; Shimada, Y.; Oberti, D.; Andriollo, A.; Lochs, S.; Bühler, M. Inheritance of a Phenotypically Neutral Epimutation Evokes Gene Silencing in Later Generations. Mol. Cell 2019, 74, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Li, A.; Calo, S.; Inoue, M.; Tonthat, N.K.; Bain, J.M.; Louw, J.; Shinohara, M.L.; Erwig, L.P.; Schumacher, M.A.; et al. Calcineurin orchestrates dimorphic transitions, antifungal drug responses and host-pathogen interactions of the pathogenic mucoralean fungus Mucor circinelloides. Mol. Microbiol. 2015, 97, 844–865. [Google Scholar] [CrossRef] [Green Version]
- Chang, Z.; Yadav, V.; Lee, S.C.; Heitman, J. Epigenetic mechanisms of drug resistance in fungi. Fungal Genet. Biol. 2019, 132, 103253. [Google Scholar] [CrossRef]
- Chang, Z.; Heitman, J. Drug-resistant epimutants exhibit organ-specific stability and induction during murine infections caused by the human fungal pathogen Mucor circinelloides. MBio 2019, 10, e02579-19. [Google Scholar] [CrossRef] [Green Version]
- Neupane, A.; Feng, C.; Mochama, P.K.; Saleem, H.; Lee Marzano, S.Y. Roles of Argonautes and Dicers on Sclerotinia sclerotiorum Antiviral RNA Silencing. Front. Plant Sci. 2019, 10, 976. [Google Scholar] [CrossRef] [Green Version]
- Nicolás, F.E.; Garre, V. RNA Interference in Fungi: Retention and Loss. In The Fungal Kingdom; Heitman, J., Howlett, B.J., Crous, P.W., Stukenbrock, E.H., James, T.Y., Gow, N.A.R., Eds.; American Society for Microbiology: Washington, DC, USA, 2017; pp. 657–672. [Google Scholar]
- Choi, J.; Kim, K.T.; Jeon, J.; Wu, J.; Song, H.; Asiegbu, F.O.; Lee, Y.H. FunRNA: A fungi-centered genomics platform for genes encoding key components of RNAi. BMC Genom. 2014, 15, S14. [Google Scholar] [CrossRef] [Green Version]
- Laurie, J.D.; Ali, S.; Linning, R.; Mannhaupt, G.; Wong, P.; Güldener, U.; Münsterkötter, M.; Moore, R.; Kahmann, R.; Bakkeren, G.; et al. Genome comparison of barley and maize smut fungi reveals targeted loss of RNA silencing components and species-specific presence of transposable elements. Plant Cell 2012, 24, 1733–1745. [Google Scholar] [CrossRef] [Green Version]
- Feretzaki, M.; Billmyre, R.B.; Clancey, S.A.; Wang, X.; Heitman, J. Gene Network Polymorphism Illuminates Loss and Retention of Novel RNAi Silencing Components in the Cryptococcus Pathogenic Species Complex. PLoS Genet. 2016, 12, e1005868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drinnenberg, I.A.; Weinberg, D.E.; Xie, K.T.; Mower, J.P.; Wolfe, K.H.; Fink, G.R.; Bartel, D.P. RNAi in budding yeast. Science 2009, 326, 544–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drinnenberg, I.A.; Fink, G.R.; Bartel, D.P. Compatibility with killer explains the rise of RNAi-deficient fungi. Science 2011, 333, 1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Eukaryotic Group | Evolutionary Advantage | Loss of RNAi | Effector Proteins | siRNAs | References |
|---|---|---|---|---|---|
| Fungi | Defense against viruses Control of TE Regulation of endogenous genes Heterochromatin formation Adaptation to stressful conditions Pathogenesis | Saccharomyces cerevisiae Ustilago maydis Cryptococcus deuterogatti | Ago Dicer Rdp/RdRP | siRNA masiRNA milRNA ex-siRNA endo-siRNA rdRNA circRNA tRF disiRNA | Table 2 and Table 3 |
| Protists | Defense against viruses Control of TE Heterochromatin formation DNA elimination | Trypanosoma cruzi Leishmania major Leishmania donovani Leishmania tarentolae Plasmodium falciparum | Piwi Dicer RdRP | siRNA scnRNA | [3,4,5,6,7] |
| Nematodes | Defense against viruses Control of TE Heterochromatin formation Regulation of endogenous genes | Ago/Piwi Dicer Rrf/Ego | miRNA piRNA endo-siRNA exo-siRNA | [3,5,8] | |
| Insects | Defense against viruses Control of TE Heterochromatin formation Regulation of endogenous genes | Ago/Piwi Dicer/Loquacious/ Drosha/Pasha/R2D2 | siRNA miRNA piRNA endo-siRNA | [3,5,8,9,10] | |
| Plants | Defense against pathogens Control of TE Heterochromatin formation Regulation of endogenous genes Repair DNA double-strand breaks (DSB) | Ago Dcl Rdr | siRNA miRNA hp-siRNA hc-siRNA or hetsiRNA tasiRNA t-siRNA lsiRNA nat-siRNA phasiRNA easiRNA diRNA | [3,8,11,12,13] | |
| Mammals | Defense against viruses Control of TE Heterochromatin formation Regulation of endogenous genes Repair DNA double-strand breaks | Ago/Piwi Dicer/Drosha | siRNA miRNA piRNA endo-siRNA diRNA | [3,5,8,13,14] |
| Function | Regulation of | Main RNAi Proteins | siRNAs | RNAi Mechanism | Fungi Species | References |
|---|---|---|---|---|---|---|
| Defense against viruses | Cryphonectria hypovirus 1 | Dcl2, Ago2 | vsRNAs | Cryphonectria parasitica | [24,25] | |
| Aspergillus virus 1816 | DclB, RsdA | vsRNAs | Aspergillus nidulans | [26] | ||
| Control of Transposable Element | LINE1-like retrotransposons | QDE-2, Dcl1, Dcl2 | Quelling | Neurospora crassa | [27] | |
| DNA transposon Sly1-1 | Dcl1, SAD-1-5, SMS-2 | masiRNAs | MSUD | Neurospora crassa | [19,28] | |
| Tf2 retrotransposon | Ago1, Rdp1, Dcr1, Clr4 | RNA-induced transcriptional silencing (RITS) | Schizosaccharomyces pombe | [29] | ||
| LTR-retrotransposon MAGGY | MoDcl2, MoAgo1, MoAgo3 | LTR-siRNAs | Magnaporthe oryzae | [30,31,32,33] | ||
| Grem-LINE1 | Ago2, Dcl1, Dcl2 | Canonical | Mucor lusitanicus | [34] | ||
| Retrotransposons | Rdp1, Dcr1, Dcr2, Ago1, Qip1, Gwc1, Srr1 | endo-siRNAs | Sex-induced silencing (SIS) Spliceosome-Coupled And Nuclear RNAi complex (SCANR) | Cryptococcus neoformans | [35,36,37,38] | |
| DNA transposon HAR1 | Rde1-5 | endo-siRNAs | Cryptococcus neoformans | [39] | ||
| Young TE | Ago1, Dicer3, RdRP1/3 | Late wave sRNAs | Puccinia graminis f. sp. triticci | [40] | ||
| Regulation of endogenous genes | RdRP1, RdRP2, Dcl1, Dcl2, Ago1, R3B2 | ex-siRNAs | Canonical | Mucor lusitanicus | [23,41] | |
| Canonical RNAi, phagocytosis response | RdRP1, RdRP3, R3B2, RnhA | rdRNAs | Non-canonical (NCRIP) | Mucor lusitanicus | [42,43] | |
| Growth and development | Dcr2, Rdr3 | ex-siRNAs | Trichoderma atroviride | [44] | ||
| Ascospore formation | FgDcl1, FgAgo2 | ex-siRNAs | Fusarium graminearum | [45] | ||
| Sexual development | FgDcl1/2, FgAgo1 | milRNAs | Fusarium graminearum | [46] | ||
| Development and fruiting body formation | milRNAs | Coprinopsis cinerea | [47] | |||
| Dicer, QIP, MRPL3, QDE-2 | milRNAs | Neurospora crassa | [48] | |||
| ERI-1, QDE-2 | disiRNAs | disiRNA loci DNA methylation (DLDM) | Neurospora crassa | [49,50] | ||
| miRNA | circRNAs | Magnaporthe oryzae | [51] | |||
| tRFs | Magnaporthe oryzae | [30] | ||||
| Plant infection | PiAgo1 | tRFs | Phytophthora infestans | [52] | ||
| Heterochromatin formation | Centromeric regions | Dcr1, Ago1, Rdp1, Tas3, Chp1 | Centromeric siRNAs | Schizosaccharomyces pombe | [53,54] | |
| Adaptation to stressful conditions | FKBP12 PyrG, PyrF | Dcr1-2, Ago1, RdRP2, QIP, RnhA | Epimutation | Mucor circinelloides Mucor lusitanicus | [55,56] | |
| Pathogenesis | Ability to infect leaves | Dcr1, Dcr2 | Colletotrichum gloeosporioides | [57] | ||
| Ability to infect leaves | Ago3, Rdp1 | Magnaportheoryzae | [58] | |||
| Virulence | Dcr1-2, Ago2 | Sclerotinia sclerotiorum | [59] | |||
| Resistance to H2O2 and virulence | Ago2 | Valsa mali | [60] | |||
| Virulence | Ago2 | Fusarium oxysporum f. sp. lycopersici | [61] | |||
| Infection | Dcr2 | Penicillium italicum | [62] | |||
| Infection | FgDcl1, FgAgo2 | Fusarium graminearum | [63] | |||
| Resistance to H2O2 and virulence | RdRP1, R3B2 | rdRNAs | NCRIP | Mucor lusitanicus | [64] | |
| Host miRNA and siRNA | PSR1 and PSR2 | Cross-kingdom RNAi | Phytophthora sojae | [65] | ||
| Host immunity genes | Bc-sRNAs | Cross-kingdom RNAi | Botrytis cinerea | [66] | ||
| Host defenses | Pst-milR1 | Cross-kingdom RNAi | Puccinia striiformis f. sp. tritici | [67] | ||
| Host metabolism genes | Bgh-sRNAs Bgt-sRNAs | Cross-kingdom RNAi | Blumeria graminis f. sp. hordei Blumeria graminis f. sp. tritici | [68] | ||
| Host genes | Cross-kingdom RNAi | Hyaloperonospora arabidopsidis | [69] |
| Species | Putative Advantage | Lost Proteins | Substitutes | References |
|---|---|---|---|---|
| Ustilago maydis | Retention of Killer virus | Ago1, RdRP1-3, Dcr1 | Partially by recombination system | [131] |
| Crytpococcus deuterogatti | Increased virulence by TE activity | Ago1, Ago2, Dcr1, Rdp1 | [96] | |
| Candida albicans | Dicer | Noncanonical CaDcr1 | [133] | |
| Naumovozyma castellii | Dicer | Noncanonical NcaDcr1 | [133] | |
| Saccharomyces cerevisiae | Retention of Killer virus | Ago1, Dcr1 | [134] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lax, C.; Tahiri, G.; Patiño-Medina, J.A.; Cánovas-Márquez, J.T.; Pérez-Ruiz, J.A.; Osorio-Concepción, M.; Navarro, E.; Calo, S. The Evolutionary Significance of RNAi in the Fungal Kingdom. Int. J. Mol. Sci. 2020, 21, 9348. https://doi.org/10.3390/ijms21249348
Lax C, Tahiri G, Patiño-Medina JA, Cánovas-Márquez JT, Pérez-Ruiz JA, Osorio-Concepción M, Navarro E, Calo S. The Evolutionary Significance of RNAi in the Fungal Kingdom. International Journal of Molecular Sciences. 2020; 21(24):9348. https://doi.org/10.3390/ijms21249348
Chicago/Turabian StyleLax, Carlos, Ghizlane Tahiri, José Alberto Patiño-Medina, José T. Cánovas-Márquez, José A. Pérez-Ruiz, Macario Osorio-Concepción, Eusebio Navarro, and Silvia Calo. 2020. "The Evolutionary Significance of RNAi in the Fungal Kingdom" International Journal of Molecular Sciences 21, no. 24: 9348. https://doi.org/10.3390/ijms21249348
APA StyleLax, C., Tahiri, G., Patiño-Medina, J. A., Cánovas-Márquez, J. T., Pérez-Ruiz, J. A., Osorio-Concepción, M., Navarro, E., & Calo, S. (2020). The Evolutionary Significance of RNAi in the Fungal Kingdom. International Journal of Molecular Sciences, 21(24), 9348. https://doi.org/10.3390/ijms21249348