Alzheimer’s, Parkinson’s Disease and Amyotrophic Lateral Sclerosis Gene Expression Patterns Divergence Reveals Different Grade of RNA Metabolism Involvement
Abstract
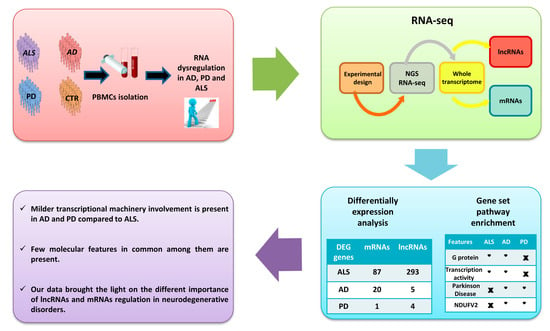
:1. Introduction
2. Results
2.1. RNA-Seq Differentially Expressed mRNAs and lncRNAs
2.1.1. Amyotrophic Lateral Sclerosis
2.1.2. Alzheimer’s Disease
2.1.3. Parkinson’s Disease
2.2. Validation of Deregulated Coding and Non-Coding Genes
2.3. mRNA Pathway Analysis
2.3.1. Amyotrophic Lateral Sclerosis
2.3.2. Alzheimer’s Disease
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. Isolation of Human Peripheral Blood Mononuclear Cells and RNA Extraction Sequencing
4.3. Libraries Preparation for RNA-Seq and Bioinformatic Data Analysis
4.4. Pathway Analysis
4.5. Real Time PCR
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SALS | Sporadic Amyotrophic Lateral Sclerosis |
| AD | Alzheimer’s disease |
| PD | Parkinson’s disease |
| CTRL | Control |
| lncRNAs | Long non-coding RNAs |
| DE | Differentially expressed |
References
- Hussain, R.; Zubair, H.; Pursell, S.; Shahab, M. Neurodegenerative Diseases: Regenerative Mechanisms and Novel Therapeutic Approaches. Brain Sci. 2018, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyss-Coray, T. Ageing, neurodegeneration and brain rejuvenation. Nature 2016, 539, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S. Molecular and Cellular Basis of Neurodegeneration in Alzheimer’s Disease. Mol. Cells 2017, 40, 613–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polymenidou, M.; Lagier-Tourenne, C.; Hutt, K.R.; Bennett, C.F.; Cleveland, D.W.; Yeo, G.W. Misregulated RNA processing in amyotrophic lateral sclerosis. Brain Res. 2012, 1462, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Serrano-Pozo, A.; Frosch, M.P.; Masliah, E.; Hyman, B.T. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2011, 1, a006189. [Google Scholar] [CrossRef]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Molteni, M.; Rossetti, C. Neurodegenerative diseases: The immunological perspective. J. Neuroimmunol. 2017, 313, 109–115. [Google Scholar] [CrossRef]
- Kiriyama, Y.; Nochi, H. The Function of Autophagy in Neurodegenerative Diseases. Int. J. Mol. Sci. 2015, 16, 26797–26812. [Google Scholar] [CrossRef] [Green Version]
- García, J.C.; Bustos, R.H. The Genetic Diagnosis of Neurodegenerative Diseases and Therapeutic Perspectives. Brain Sci. 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Liu, E.Y.; Cali, C.P.; Lee, E.B. RNA metabolism in neurodegenerative disease. Dis. Model. Mech. 2017, 10, 509–518. [Google Scholar] [CrossRef] [Green Version]
- Alaqeel, A.M.; Abou Al-Shaar, H.; Shariff, R.K.; Albakr, A. The role of RNA metabolism in neurological diseases. Balk. J. Med. Genet. 2015, 18, 5–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weskamp, K.; Barmada, S.J. RNA Degradation in Neurodegenerative Disease. Adv. Neurobiol. 2018, 20, 103–142. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Romero, C.; Hur, J.; Lunn, J.S.; Paez-Colasante, X.; Bender, D.E.; Yung, R.; Sakowski, S.A.; Feldman, E.L. Expression of microRNAs in human post-mortem amyotrophic lateral sclerosis spinal cords provides insight into disease mechanisms. Mol. Cell. Neurosci. 2016, 71, 34–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Wei, Q.; Chen, X.; Li, C.; Cao, B.; Ou, R.; Hadano, S.; Shang, H.F. Aberration of miRNAs Expression in Leukocytes from Sporadic Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2016, 9, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimoto, Y.; Nakagawa, S.; Hirose, T.; Okano, H.J.; Takao, M.; Shibata, S.; Suyama, S.; Kuwako, K.; Imai, T.; Murayama, S.; et al. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 2013, 6, 31. [Google Scholar] [CrossRef] [Green Version]
- Sunwoo, J.S.; Lee, S.T.; Im, W.; Lee, M.; Byun, J.I.; Jung, K.H.; Park, K.I.; Jung, K.Y.; Lee, S.K.; Chu, K.; et al. Altered Expression of the Long Noncoding RNA NEAT1 in Huntington’s Disease. Mol. Neurobiol. 2017, 54, 1577–1586. [Google Scholar] [CrossRef]
- Simchovitz, A.; Hanan, M.; Niederhoffer, N.; Madrer, N.; Yayon, N.; Bennett, E.R.; Greenberg, D.S.; Kadener, S.; Soreq, H. NEAT1 is overexpressed in Parkinson’s disease substantia nigra and confers drug-inducible neuroprotection from oxidative stress. FASEB J. 2019, 33, 11223–11234. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Zhang, Y.; Wang, S.; Yang, F.; Cai, H.; Xing, Y.; Chen, Z.; Chen, J. NEAT1 Decreasing Suppresses Parkinson’s Disease Progression via Acting as miR-1301-3p Sponge. J. Mol. Neurosci. 2020. [Google Scholar] [CrossRef]
- Kapeli, K.; Martinez, F.J.; Yeo, G.W. Genetic mutations in RNA-binding proteins and their roles in ALS. Hum. Genet 2017, 136, 1193–1214. [Google Scholar] [CrossRef] [Green Version]
- Millan, M.J. Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer’s disease: An integrative review. Prog. Neurobiol. 2017, 156, 1–68. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Sen, D. MicroRNAs in Parkinson’s disease. Exp. Brain Res. 2017, 235, 2359–2374. [Google Scholar] [CrossRef]
- Byron, S.A.; van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet 2016, 17, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, S.; Zucca, S.; Pandini, C.; Diamanti, L.; Bordoni, M.; Sproviero, D.; Arigoni, M.; Olivero, M.; Pansarasa, O.; Ceroni, M.; et al. Long non-coding and coding RNAs characterization in Peripheral Blood Mononuclear Cells and Spinal Cord from Amyotrophic Lateral Sclerosis patients. Sci. Rep. 2018, 8, 2378. [Google Scholar] [CrossRef]
- Luotti, S.; Pasetto, L.; Porcu, L.; Torri, V.; Elezgarai, S.R.; Pantalone, S.; Filareti, M.; Corbo, M.; Lunetta, C.; Mora, G.; et al. Diagnostic and prognostic values of PBMC proteins in amyotrophic lateral sclerosis. Neurobiol. Dis. 2020, 139, 104815. [Google Scholar] [CrossRef] [PubMed]
- Mosallaei, M.; Ehtesham, N.; Rahimirad, S.; Saghi, M.; Vatandoost, N.; Khosravi, S. PBMCs: A new source of diagnostic and prognostic biomarkers. Arch. Physiol. Biochem. 2020, 1–7. [Google Scholar] [CrossRef]
- Gagliardi, S.; Franco, V.; Sorrentino, S.; Zucca, S.; Pandini, C.; Rota, P.; Bernuzzi, S.; Costa, A.; Sinforiani, E.; Pansarasa, O.; et al. Curcumin and Novel Synthetic Analogs in Cell-Based Studies of Alzheimer’s Disease. Front. Pharmacol. 2018, 9, 1404. [Google Scholar] [CrossRef]
- Lazo-Gómez, R.; Ramírez-Jarquín, U.N.; Tovar-Y-Romo, L.B.; Tapia, R. Histone deacetylases and their role in motor neuron degeneration. Front. Cell. Neurosci. 2013, 7, 243. [Google Scholar] [CrossRef]
- Kim, T.; Cui, R.; Jeon, Y.J.; Fadda, P.; Alder, H.; Croce, C.M. MYC-repressed long noncoding RNAs antagonize MYC-induced cell proliferation and cell cycle progression. Oncotarget 2015, 6, 18780–18789. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Qi, X. VCP cooperates with UBXD1 to degrade mitochondrial outer membrane protein MCL1 in model of Huntington’s disease. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 552–559. [Google Scholar] [CrossRef]
- Lee, J.K.; Shin, J.H.; Lee, J.E.; Choi, E.J. Role of autophagy in the pathogenesis of amyotrophic lateral sclerosis. Biochim. Biophys. Acta 2015, 1852, 2517–2524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villar-Menéndez, I.; Porta, S.; Buira, S.P.; Pereira-Veiga, T.; Díaz-Sánchez, S.; Albasanz, J.L.; Ferrer, I.; Martín, M.; Barrachina, M. Increased striatal adenosine A2A receptor levels is an early event in Parkinson’s disease-related pathology and it is potentially regulated by miR-34b. Neurobiol. Dis. 2014, 69, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Castillo, E.; Leon, J.; Mazzei, G.; Abolhassani, N.; Haruyama, N.; Saito, T.; Saido, T.; Hokama, M.; Iwaki, T.; Ohara, T.; et al. Comparative profiling of cortical gene expression in Alzheimer’s disease patients and mouse models demonstrates a link between amyloidosis and neuroinflammation. Sci. Rep. 2017, 7, 17762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, Y.; Niikura, T.; Tajima, H.; Yasukawa, T.; Sudo, H.; Ito, Y.; Kita, Y.; Kawasumi, M.; Kouyama, K.; Doyu, M.; et al. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Abeta. Proc. Natl. Acad. Sci. USA 2001, 98, 6336–6341. [Google Scholar] [CrossRef] [Green Version]
- Grace, E.A.; Busciglio, J. Aberrant activation of focal adhesion proteins mediates fibrillar amyloid beta-induced neuronal dystrophy. J. Neurosci. 2003, 23, 493–502. [Google Scholar] [CrossRef] [Green Version]
- Calderwood, D.A.; Campbell, I.D.; Critchley, D.R. Talins and kindlins: Partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 2013, 14, 503–517. [Google Scholar] [CrossRef] [Green Version]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef]
- Ju, X.C.; Hou, Q.Q.; Sheng, A.L.; Wu, K.Y.; Zhou, Y.; Jin, Y.; Wen, T.; Yang, Z.; Wang, X.; Luo, Z.G. The hominoid-specific gene TBC1D3 promotes generation of basal neural progenitors and induces cortical folding in mice. Elife 2016, 5. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Nishioka, K.; Vilariño-Güell, C.; Cobb, S.A.; Kachergus, J.M.; Ross, O.A.; Hentati, E.; Hentati, F.; Farrer, M.J. Genetic variation of the mitochondrial complex I subunit NDUFV2 and Parkinson’s disease. Parkinsonism Relat. Disord. 2010, 16, 686–687. [Google Scholar] [CrossRef] [Green Version]
- Troncone, L.; Luciani, M.; Coggins, M.; Wilker, E.H.; Ho, C.Y.; Codispoti, K.E.; Frosch, M.P.; Kayed, R.; Del Monte, F. Aβ Amyloid Pathology Affects the Hearts of Patients With Alzheimer’s Disease: Mind the Heart. J. Am. Coll. Cardiol. 2016, 68, 2395–2407. [Google Scholar] [CrossRef] [PubMed]
- Gdynia, H.J.; Kurt, A.; Endruhn, S.; Ludolph, A.C.; Sperfeld, A.D. Cardiomyopathy in motor neuron diseases. J. Neurol. Neurosurg. Psychiatry 2006, 77, 671–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orsini, F.; de Blasio, D.; Zangari, R.; Zanier, E.R.; de Simoni, M.G. Versatility of the complement system in neuroinflammation, neurodegeneration and brain homeostasis. Front. Cell. Neurosci. 2014, 8, 380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasuri, K.; Zhang, L.; Keller, J.N. Oxidative stress, neurodegeneration, and the balance of protein degradation and protein synthesis. Free. Radic. Biol. Med. 2013, 62, 170–185. [Google Scholar] [CrossRef]
- Kim, G.H.; Kim, J.E.; Rhie, S.J.; Yoon, S. The Role of Oxidative Stress in Neurodegenerative Diseases. Exp. Neurobiol. 2015, 24, 325–340. [Google Scholar] [CrossRef]
- Wei, C.W.; Luo, T.; Zou, S.S.; Wu, A.S. The Role of Long Noncoding RNAs in Central Nervous System and Neurodegenerative Diseases. Front. Behav. Neurosci. 2018, 12, 175. [Google Scholar] [CrossRef]
- Soreq, L.; Guffanti, A.; Salomonis, N.; Simchovitz, A.; Israel, Z.; Bergman, H.; Soreq, H. Long non-coding RNA and alternative splicing modulations in Parkinson’s leukocytes identified by RNA sequencing. PLoS Comput. Biol. 2014, 10, e1003517. [Google Scholar] [CrossRef]
- Elkouris, M.; Kouroupi, G.; Vourvoukelis, A.; Papagiannakis, N.; Kaltezioti, V.; Matsas, R.; Stefanis, L.; Xilouri, M.; Politis, P.K. Long Non-coding RNAs Associated With Neurodegeneration-Linked Genes Are Reduced in Parkinson’s Disease Patients. Front. Cell. Neurosci. 2019, 13, 58. [Google Scholar] [CrossRef] [Green Version]
- Gérard, M.A.; Myslinski, E.; Chylak, N.; Baudrey, S.; Krol, A.; Carbon, P. The scaRNA2 is produced by an independent transcription unit and its processing is directed by the encoding region. Nucleic Acids Res. 2010, 38, 370–381. [Google Scholar] [CrossRef] [Green Version]
- Brooks, B.R. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J. Neurol. Sci. 1994, 124, 96–107. [Google Scholar] [CrossRef]
- McKhann, G.; Drachman, D.; Folstein, M.; Katzman, R.; Price, D.; Stadlan, E.M. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984, 34, 939–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Postuma, R.B.; Berg, D.; Stern, M.; Poewe, W.; Olanow, C.W.; Oertel, W.; Obeso, J.; Marek, K.; Litvan, I.; Lang, A.E.; et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov. Disord. 2015, 30, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrara, M.; Lum, J.; Cordero, F.; Beccuti, M.; Poidinger, M.; Donatelli, S.; Calogero, R.A.; Zolezzi, F. Alternative splicing detection workflow needs a careful combination of sample prep and bioinformatics analysis. BMC Bioinform. 2015, 16 (Suppl. 9), S2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]









| SALS | AD | PD | ||||
|---|---|---|---|---|---|---|
| mRNA | lncRNA | mRNA | lncRNA | mRNA | lncRNA | |
| Up-regulated | 57 | 183 | 8 | 3 | 0 | 1 |
| Down-regulated | 30 | 110 | 11 | 1 | 1 | 3 |
| Total | 87 | 293 | 19 | 4 | 1 | 4 |
| KEGG/GO Term | SALS | AD |
|---|---|---|
| Dilated cardiomyopathy (DCM) | ADCY9 PLN | ITGA2B |
| Complement and coagulation cascades | A2M F2RL2 | F13A1 |
| Fluid shear stress and atherosclerosis | RELA ACVR2A NFE2L2 | NCF1 ITGA2B |
| Cellular response to reactive oxygen species | MPV17L RELA NFE2L2 | NCF1 |
| Protein localization cell surface | SMURF1 | VCL |
| SALS | AD | PD | CTRL | |
|---|---|---|---|---|
| n = 10 | n = 6 | n = 6 | n = 14 | |
| Age (M ± SD) | 66.6 ± 10.1 | 78.2 ± 8.0 | 62.7 ± 6.7 | 53.2 ± 8.3 |
| Sex | ||||
| Males n (%) | 45% | 34% | 83% | 64% |
| Females n (%) | 55% | 66% | 17% | 32% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garofalo, M.; Pandini, C.; Bordoni, M.; Pansarasa, O.; Rey, F.; Costa, A.; Minafra, B.; Diamanti, L.; Zucca, S.; Carelli, S.; et al. Alzheimer’s, Parkinson’s Disease and Amyotrophic Lateral Sclerosis Gene Expression Patterns Divergence Reveals Different Grade of RNA Metabolism Involvement. Int. J. Mol. Sci. 2020, 21, 9500. https://doi.org/10.3390/ijms21249500
Garofalo M, Pandini C, Bordoni M, Pansarasa O, Rey F, Costa A, Minafra B, Diamanti L, Zucca S, Carelli S, et al. Alzheimer’s, Parkinson’s Disease and Amyotrophic Lateral Sclerosis Gene Expression Patterns Divergence Reveals Different Grade of RNA Metabolism Involvement. International Journal of Molecular Sciences. 2020; 21(24):9500. https://doi.org/10.3390/ijms21249500
Chicago/Turabian StyleGarofalo, Maria, Cecilia Pandini, Matteo Bordoni, Orietta Pansarasa, Federica Rey, Alfredo Costa, Brigida Minafra, Luca Diamanti, Susanna Zucca, Stephana Carelli, and et al. 2020. "Alzheimer’s, Parkinson’s Disease and Amyotrophic Lateral Sclerosis Gene Expression Patterns Divergence Reveals Different Grade of RNA Metabolism Involvement" International Journal of Molecular Sciences 21, no. 24: 9500. https://doi.org/10.3390/ijms21249500
APA StyleGarofalo, M., Pandini, C., Bordoni, M., Pansarasa, O., Rey, F., Costa, A., Minafra, B., Diamanti, L., Zucca, S., Carelli, S., Cereda, C., & Gagliardi, S. (2020). Alzheimer’s, Parkinson’s Disease and Amyotrophic Lateral Sclerosis Gene Expression Patterns Divergence Reveals Different Grade of RNA Metabolism Involvement. International Journal of Molecular Sciences, 21(24), 9500. https://doi.org/10.3390/ijms21249500