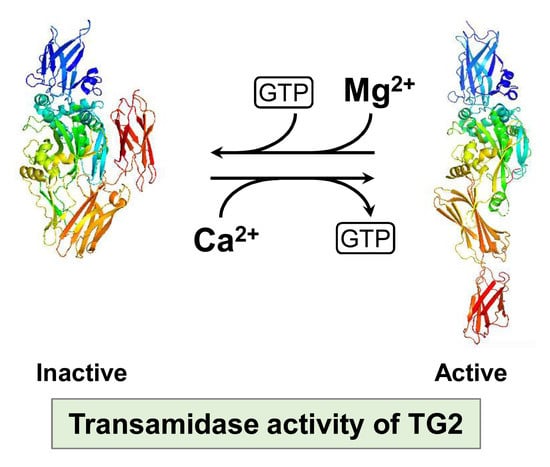
Competitive Binding of Magnesium to Calcium Binding Sites Reciprocally Regulates Transamidase and GTP Hydrolysis Activity of Transglutaminase 2
Abstract
:1. Introduction
2. Results
2.1. Identification of New Ca2+-Binding Sites in TG2
2.2. Ca2+ Binding to E437 and E539 Residues Is Required for the Transamidase Activity of TG2
2.3. Mg2+ Binding to E437 and E539 Inhibits the Transamidase Activity of TG2
2.4. Mg2+-Binding to E437 and E539 Promotes the GTP Binding and Hydrolysis Activity of TG2
2.5. Mg2+-Binding to E437 and E539 Is Critical for Preventing the Activation of Intracellular TG2
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Crystallization and Data Collection
4.3. Structure Determination and Analysis
4.4. CD Analysis
4.5. Protein Data Bank Accession Code
4.6. ITC
4.7. In Vitro Transamidase Activity Assay
4.8. GTP Binding Assay
4.9. GTPase Activity Assay
4.10. Cell Culture
4.11. Intracellular Transamidase Activity Assay
4.12. Luciferase Reporter Assay
4.13. Western Blot Analysis
4.14. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TG2 | Transglutaminase 2 |
| Ca2+ | Calcium ion |
| Mg2+ | Magnesium ion |
| ASU | Asymmetric unit |
| CD | Circular dichroism |
| WT | Wild type |
| ITC | Isothermal titration calorimetry |
| EC50 | Half-maximal effective concentration |
| IC50 | Half-maximal inhibitory concentration |
| NB | No binding |
| HPSP | Human phosphoserine phosphatase |
| IPTG | Isopropyl β-D-thiogalactopyranoside |
| BP | Biotinylated pentylamine |
| HDF | Human dermal fibroblasts |
| CI | Confidence intervals |
| n.s. | Not significant |
References
- Tatsukawa, H.; Furutani, Y.; Hitomi, K.; Kojima, S. Transglutaminase 2 has opposing roles in the regulation of cellular functions as well as cell growth and death. Cell Death Dis. 2016, 7, e2244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorand, L.; Graham, R.M. Transglutaminases: Crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 2003, 4, 140–156. [Google Scholar] [CrossRef] [PubMed]
- Iismaa, S.E.; Mearns, B.M.; Lorand, L.; Graham, R.M. Transglutaminases and disease: Lessons from genetically engineered mouse models and inherited disorders. Physiol. Rev. 2009, 89, 991–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, D.M.; Jeon, J.H.; Kim, C.W.; Cho, S.Y.; Kwon, J.C.; Lee, H.J.; Choi, K.H.; Park, S.C.; Kim, I.G. Cell type-specific activation of intracellular transglutaminase 2 by oxidative stress or ultraviolet irradiation: Implications of transglutaminase 2 in age-related cataractogenesis. J. Biol. Chem. 2004, 279, 15032–15039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, E.M.; Kim, C.W.; Cho, S.Y.; Jang, G.Y.; Shin, D.M.; Jeon, J.H.; Kim, I.G. Degradation of transglutaminase 2 by calcium-mediated ubiquitination responding to high oxidative stress. FEBS Lett. 2009, 583, 648–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.Y.; Jeong, E.M.; Lee, J.H.; Kim, H.J.; Lim, J.; Kim, C.W.; Shin, D.M.; Jeon, J.H.; Choi, K.; Kim, I.G. Doxorubicin induces the persistent activation of intracellular transglutaminase 2 that protects from cell death. Mol. Cells 2012, 33, 235–241. [Google Scholar] [CrossRef] [Green Version]
- Oh, K.; Park, H.B.; Byoun, O.J.; Shin, D.M.; Jeong, E.M.; Kim, Y.W.; Kim, Y.S.; Melino, G.; Kim, I.G.; Lee, D.S. Epithelial transglutaminase 2 is needed for T cell interleukin-17 production and subsequent pulmonary inflammation and fibrosis in bleomycin-treated mice. J. Exp. Med. 2011, 208, 1707–1719. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Cerione, R.A.; Clardy, J. Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity. Proc. Natl. Acad. Sci. USA 2002, 99, 2743–2747. [Google Scholar] [CrossRef] [Green Version]
- Jang, T.H.; Lee, D.S.; Choi, K.; Jeong, E.M.; Kim, I.G.; Kim, Y.W.; Chun, J.N.; Jeon, J.H.; Park, H.H. Crystal structure of transglutaminase 2 with GTP complex and amino acid sequence evidence of evolution of GTP binding site. PLoS ONE 2014, 9, e107005. [Google Scholar] [CrossRef] [Green Version]
- Pinkas, D.M.; Strop, P.; Brunger, A.T.; Khosla, C. Transglutaminase 2 undergoes a large conformational change upon activation. PLoS Biol. 2007, 5, e327. [Google Scholar] [CrossRef]
- Bergamini, C.M. GTP modulates calcium binding and cation-induced conformational changes in erythrocyte transglutaminase. FEBS Lett. 1988, 239, 255–258. [Google Scholar] [CrossRef] [Green Version]
- Kiraly, R.; Csosz, E.; Kurtan, T.; Antus, S.; Szigeti, K.; Simon-Vecsei, Z.; Korponay-Szabo, I.R.; Keresztessy, Z.; Fesus, L. Functional significance of five noncanonical Ca2+-binding sites of human transglutaminase 2 characterized by site-directed mutagenesis. FEBS J. 2009, 276, 7083–7096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, B.A.; Yee, V.C.; Pedersen, L.C.; Le Trong, I.; Bishop, P.D.; Stenkamp, R.E.; Teller, D.C. Identification of the calcium binding site and a novel ytterbium site in blood coagulation factor XIII by x-ray crystallography. J. Biol. Chem. 1999, 274, 4917–4923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stieler, M.; Weber, J.; Hils, M.; Kolb, P.; Heine, A.; Buchold, C.; Pasternack, R.; Klebe, G. Structure of active coagulation factor XIII triggered by calcium binding: Basis for the design of next-generation anticoagulants. Angew. Chem. Int. Ed. Engl. 2013, 52, 11930–11934. [Google Scholar] [CrossRef] [PubMed]
- Ahvazi, B.; Kim, H.C.; Kee, S.H.; Nemes, Z.; Steinert, P.M. Three-dimensional structure of the human transglutaminase 3 enzyme: Binding of calcium ions changes structure for activation. EMBO J. 2002, 21, 2055–2067. [Google Scholar] [CrossRef] [PubMed]
- Romani, A.M. Magnesium homeostasis in mammalian cells. Front. Biosci. 2007, 12, 308–331. [Google Scholar] [CrossRef]
- Barbagallo, M.; Belvedere, M.; Dominguez, L.J. Magnesium homeostasis and aging. Magnes. Res. 2009, 22, 235–246. [Google Scholar] [CrossRef] [Green Version]
- Clapham, D.E. Calcium signaling. Cell 2007, 131, 1047–1058. [Google Scholar] [CrossRef] [Green Version]
- Killilea, D.W.; Ames, B.N. Magnesium deficiency accelerates cellular senescence in cultured human fibroblasts. Proc. Natl. Acad. Sci. USA 2008, 105, 5768–5773. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, J.; Romani, A.M.; Valentin-Torres, A.M.; Luciano, A.A.; Ramirez Kitchen, C.M.; Funderburg, N.; Mesiano, S.; Bernstein, H.B. Magnesium decreases inflammatory cytokine production: A novel innate immunomodulatory mechanism. J. Immunol. 2012, 188, 6338–6346. [Google Scholar] [CrossRef] [Green Version]
- Elsasser, H.P.; MacDonald, R.; Dienst, M.; Kern, H.F. Characterization of a transglutaminase expressed in human pancreatic adenocarcinoma cells. Eur J Cell Biol 1993, 61, 321–328. [Google Scholar]
- Lai, T.S.; Slaughter, T.F.; Peoples, K.A.; Hettasch, J.M.; Greenberg, C.S. Regulation of human tissue transglutaminase function by magnesium-nucleotide complexes. Identification of distinct binding sites for Mg-GTP and Mg-ATP. J. Biol. Chem. 1998, 273, 1776–1781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birnbaumer, L.; Zurita, A.R. On the roles of Mg in the activation of G proteins. J. Recept Signal Transduct. Res. 2010, 30, 372–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sprang, S.R. Invited review: Activation of G proteins by GTP and the mechanism of Galpha-catalyzed GTP hydrolysis. Biopolymers 2016, 105, 449–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, T.H.; Park, H.H. Crystallization and preliminary X-ray crystallographic studies of transglutaminase 2 in complex with Ca2+. Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 513–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahvazi, B.; Boeshans, K.M.; Idler, W.; Baxa, U.; Steinert, P.M. Roles of calcium ions in the activation and activity of the transglutaminase 3 enzyme. J. Biol. Chem. 2003, 278, 23834–23841. [Google Scholar] [CrossRef] [Green Version]
- Begg, G.E.; Holman, S.R.; Stokes, P.H.; Matthews, J.M.; Graham, R.M.; Iismaa, S.E. Mutation of a critical arginine in the GTP-binding site of transglutaminase 2 disinhibits intracellular cross-linking activity. J. Biol. Chem. 2006, 281, 12603–12609. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Lee, K.B.; Son, Y.H.; Shin, J.; Lee, J.H.; Kim, H.J.; Hong, A.Y.; Bae, H.W.; Kwon, M.A.; Lee, W.J.; et al. Transglutaminase 2 mediates UV-induced skin inflammation by enhancing inflammatory cytokine production. Cell Death. Dis. 2017, 8, e3148. [Google Scholar] [CrossRef]
- Peeraer, Y.; Rabijns, A.; Collet, J.F.; Van Schaftingen, E.; De Ranter, C. How calcium inhibits the magnesium-dependent enzyme human phosphoserine phosphatase. Eur. J. Biochem. 2004, 271, 3421–3427. [Google Scholar] [CrossRef]
- Traut, T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 1994, 140, 1–22. [Google Scholar] [CrossRef]
- Rudack, T.; Xia, F.; Schlitter, J.; Kotting, C.; Gerwert, K. The role of magnesium for geometry and charge in GTP hydrolysis, revealed by quantum mechanics/molecular mechanics simulations. Biophys. J. 2012, 103, 293–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shutes, A.; Phillips, R.A.; Corrie, J.E.; Webb, M.R. Role of magnesium in nucleotide exchange on the small G protein rac investigated using novel fluorescent Guanine nucleotide analogues. Biochemistry 2002, 41, 3828–3835. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Shrubsole, M.J.; Ness, R.M.; Schlundt, D.; Cai, Q.; Smalley, W.E.; Li, M.; Shyr, Y.; Zheng, W. The relation of magnesium and calcium intakes and a genetic polymorphism in the magnesium transporter to colorectal neoplasia risk. Am. J. Clin. Nutr. 2007, 86, 743–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorczyca, A.M.; He, K.; Xun, P.; Margolis, K.L.; Wallace, J.P.; Lane, D.; Thomson, C.; Ho, G.Y.; Shikany, J.M.; Luo, J. Association between magnesium intake and risk of colorectal cancer among postmenopausal women. Cancer Causes Control 2015, 26, 1761–1769. [Google Scholar] [CrossRef]
- Larsson, S.C.; Bergkvist, L.; Wolk, A. Magnesium intake in relation to risk of colorectal cancer in women. JAMA 2005, 293, 86–89. [Google Scholar] [CrossRef] [Green Version]
- Dibaba, D.; Xun, P.; Yokota, K.; White, E.; He, K. Magnesium intake and incidence of pancreatic cancer: The VITamins and Lifestyle study. Br. J. Cancer 2015, 113, 1615–1621. [Google Scholar] [CrossRef]
- Hruby, A.; O’Donnell, C.J.; Jacques, P.F.; Meigs, J.B.; Hoffmann, U.; McKeown, N.M. Magnesium intake is inversely associated with coronary artery calcification: The Framingham Heart Study. JACC Cardiovasc. Imaging 2014, 7, 59–69. [Google Scholar] [CrossRef] [Green Version]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- McCoy, A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007, 63, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLano, W.L.; Lam, J.W. PyMOL: A communications tool for computational models. Abstr. Pap. Am. Chem. S 2005, 230, U1371–U1372. [Google Scholar]
- Ha, H.J.; Kwon, S.; Jeong, E.M.; Kim, C.M.; Lee, K.B.; Kim, I.G.; Park, H.H. Structure of natural variant transglutaminase 2 reveals molecular basis of gaining stability and higher activity. PLoS ONE 2018, 13, e0204707. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.Y.; Jeon, J.H.; Cho, S.Y.; Shin, D.M.; Kim, C.W.; Jeong, E.M.; Bae, H.C.; Kim, T.W.; Lee, S.H.; Choi, Y.; et al. Transglutaminase 2 suppresses apoptosis by modulating caspase 3 and NF-kappaB activity in hypoxic tumor cells. Oncogene 2010, 29, 356–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Data Collection | Native |
|---|---|
| X-ray source | Synchrotron (PAL 5C) |
| Detector | Eiger 9M |
| Wavelength | 1.0000 |
| Space group | C2221 |
| Cell dimensions | |
| a, b, c | 133.1 Å, 216.3 Å, 166.3 Å |
| Resolution | 50–3.56 Å |
| Wilson B-factor | 80.666 Å2 |
| †No. of unique reflections overall | 29,265 |
| † Rsym | 9.4% (34.5%) |
| †I/I | 19.2 (3.9) |
| †Completeness | 100% (99.9%) |
| †Redundancy | 10.9 (11.0) |
| Refinement | |
| Resolution | 42.58–3.55 Å |
| No. of reflections used (completeness) | 27,758 (99.59%) |
| †Rwork | 22.5% (22.68%) |
| †Rfree | 26.3% (26.85%) |
| Average B-factors | |
| Protein | 61.0 Å2 |
| Other small molecules | 82 Å2 |
| Root mean square deviations | |
| Bond lengths | 0.013 Å |
| Bond angles | 1.672° |
| MolProbity analysis | |
| Ramachandran outliers | 0.00% |
| Ramachandran favored | 98.00% |
| Ramachandran allowed | 2.00% |
| Rotamer outliers | 6.00% |
| Clashscore | 18.00 |
| Divalent Ions | WT Kd (μM) | E437R Kd (μM) | E539R Kd (μM) | E437R/E539R Kd (μM) |
|---|---|---|---|---|
| Ca2+ | 0.027 ± 0.004 | 188 ± 24 | 0.047 ± 0.001 | NB |
| Mg2+ | 0.215 ± 0.056 | 1.459 ± 0.523 | 0.282 ± 0.023 | NB |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, E.M.; Lee, K.B.; Kim, G.E.; Kim, C.M.; Lee, J.-H.; Kim, H.-J.; Shin, J.-W.; Kwon, M.-a.; Park, H.H.; Kim, I.-G. Competitive Binding of Magnesium to Calcium Binding Sites Reciprocally Regulates Transamidase and GTP Hydrolysis Activity of Transglutaminase 2. Int. J. Mol. Sci. 2020, 21, 791. https://doi.org/10.3390/ijms21030791
Jeong EM, Lee KB, Kim GE, Kim CM, Lee J-H, Kim H-J, Shin J-W, Kwon M-a, Park HH, Kim I-G. Competitive Binding of Magnesium to Calcium Binding Sites Reciprocally Regulates Transamidase and GTP Hydrolysis Activity of Transglutaminase 2. International Journal of Molecular Sciences. 2020; 21(3):791. https://doi.org/10.3390/ijms21030791
Chicago/Turabian StyleJeong, Eui Man, Ki Baek Lee, Gi Eob Kim, Chang Min Kim, Jin-Haeng Lee, Hyo-Jun Kim, Ji-Woong Shin, Mee-ae Kwon, Hyun Ho Park, and In-Gyu Kim. 2020. "Competitive Binding of Magnesium to Calcium Binding Sites Reciprocally Regulates Transamidase and GTP Hydrolysis Activity of Transglutaminase 2" International Journal of Molecular Sciences 21, no. 3: 791. https://doi.org/10.3390/ijms21030791
APA StyleJeong, E. M., Lee, K. B., Kim, G. E., Kim, C. M., Lee, J. -H., Kim, H. -J., Shin, J. -W., Kwon, M. -a., Park, H. H., & Kim, I. -G. (2020). Competitive Binding of Magnesium to Calcium Binding Sites Reciprocally Regulates Transamidase and GTP Hydrolysis Activity of Transglutaminase 2. International Journal of Molecular Sciences, 21(3), 791. https://doi.org/10.3390/ijms21030791