Cellular Localization of Carbonic Anhydrase Nce103p in Candida albicans and Candida parapsilosis
Abstract
:1. Introduction
2. Results
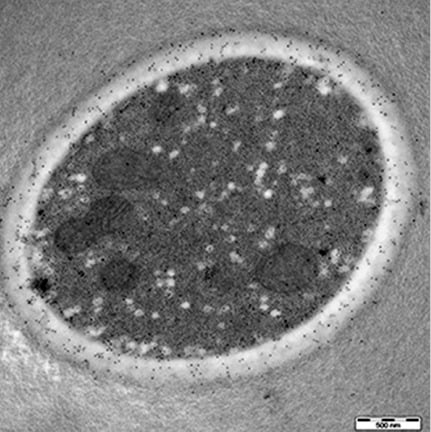
2.1. Localization of Candida CAs Using Immunogold Labeling and Electron Microscopy
2.2. Mass-Spectrometry Analysis of CW and PM Fractions
2.3. Western Blot Analysis of Selected Subcellular Fractions
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Immunogold Labeling and Electron Microscopy
4.3. Isolation of the Cell Wall
4.4. Mass Spectrometry Analysis of Nce103p in the Cell Wall Samples
4.5. Preparation of Spheroplasts and Subcellular Fractions
4.6. Western Blot Analysis of Subcellular Fractions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guinea, J. Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 2014, 8, 5–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Ami, R. Treatment of invasive candidiasis: A narrative review. J. Fungi 2018, 4, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tóth, R.; Nosek, J.; Mora-Montes, H.M.; Gabaldon, T.; Bliss, J.M.; Nosanchuk, J.D.; Turner, S.A.; Butler, G.; Vágvölgyi, C.; Gácser, A. Candida parapsilosis: From Genes to the Bedside. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klengel, T.; Liang, W.-J.J.; Chaloupka, J.; Ruoff, C.; Schröppel, K.; Naglik, J.R.; Eckert, S.E.; Mogensen, E.G.; Haynes, K.; Tuite, M.F.; et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 2005, 15, 2021–2026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segal, E.S.; Gritsenko, V.; Levitan, A.; Yadav, B.; Dror, N.; Steenwyk, J.L.; Silberberg, Y.; Mielich, K.; Rokas, A.; Gow, N.A.R.; et al. Gene Essentiality Analyzed by In Vivo Transposon Mutagenesis and Machine Learning in a Stable Haploid Isolate of Candida albicans. MBio 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cottier, F.; Raymond, M.; Kurzai, O.; Bolstad, M.; Leewattanapasuk, W.; Jiménez-López, C.; Lorenz, M.C.; Sanglard, D.; Váchová, L.; Pavelka, N.; et al. The bZIP transcription factor Rca1p is a central regulator of a novel CO 2 sensing pathway in yeast. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, R.; Pohlers, S.; Mühlschlegel, F.A.; Kurzai, O. CO2 sensing in fungi: At the heart of metabolic signaling. Curr. Genet. 2017, 63, 965–972. [Google Scholar] [CrossRef] [PubMed]
- Elleuche, S.; Pöggeler, S. Carbonic anhydrases in fungi. Microbiology 2010, 156, 23–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, Y.-B.; Jiang, Y.-L.; He, Y.-X.; He, W.-W.; Lian, F.-M.; Chen, Y.; Zhou, C.-Z. Structural insights into the substrate tunnel of Saccharomyces cerevisiae carbonic anhydrase Nce103. BMC Struct. Biol. 2009, 9, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dostál, J.; Brynda, J.; Blaha, J.; Macháček, S.; Heidingsfeld, O.; Pichová, I. Crystal structure of carbonic anhydrase CaNce103p from the pathogenic yeast Candida albicans. BMC Struct. Biol. 2018, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Götz, R.; Gnann, A.; Zimmermann, F.K. Deletion of the carbonic anhydrase-like geneNCE103 of the yeastSaccharomyces cerevisiae causes an oxygen-sensitive growth defect. Yeast 1999, 15, 855–864. [Google Scholar] [CrossRef]
- Cleves, A.E.; Cooper, D.N.W.; Barondes, S.H.; Kelly, R.B. A new pathway for protein export in Saccharomyces cerevisiae. J. Cell Biol. 1996, 133, 1017–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skrzypek, M.; Binkley, J.; Binkley, G.; Miyasato, S.; Simison, M.; Sherlock, G. Candida Genome Database. Available online: http://www.candidagenome.org/ (accessed on 20 January 2020).
- Zinser, E.; Daum, G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast 1995, 11, 493–536. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Klis, F.M.; Brul, S. Adaptations of the secretome of Candida albicans in response to host-related environmental conditions. Eukaryot. Cell 2015, 14, 1165–1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- She, X.; Zhang, P.; Gao, Y.; Zhang, L.; Wang, Q.; Chen, H.; Calderone, R.; Liu, W.; Li, D. A mitochondrial proteomics view of complex I deficiency in Candida albicans. Mitochondrion 2018, 38, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Vögtle, F.N.; Burkhart, J.M.; Rao, S.; Gerbeth, C.; Hinrichs, J.; Martinou, J.C.; Chacinska, A.; Sickmann, A.; Zahedi, R.P.; Meisinger, C. Intermembrane space proteome of yeast mitochondria. Mol. Cell. Proteomics 2012, 11, 1840–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Agarwal, S.; Heyman, J.A.; Matson, S.; Heidtman, M.; Piccirillo, S.; Umansky, L.; Drawid, A.; Jansen, R.; Liu, Y.; et al. Subcellular localization of the yeast proteome. Genes Dev. 2002, 16, 707–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitarch, A.; Sánchez, M.; Nombela, C.; Gil, C. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol. Cell. Proteomics 2002, 1, 967–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinterová, Z.; Šanda, M.; Dostál, J.; Hrušková-Heidingsfeldová, O.; Pichová, I. Evidence for the presence of proteolytically active secreted aspartic proteinase 1 of Candida parapsilosis in the cell wall. Protein Sci. 2011, 20, 2004–2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dostál, J.; Blaha, J.; Hadravová, R.; Hubálek, M.; Heidingsfeld, O.; Pichová, I. Cellular Localization of Carbonic Anhydrase Nce103p in Candida albicans and Candida parapsilosis. Int. J. Mol. Sci. 2020, 21, 850. https://doi.org/10.3390/ijms21030850
Dostál J, Blaha J, Hadravová R, Hubálek M, Heidingsfeld O, Pichová I. Cellular Localization of Carbonic Anhydrase Nce103p in Candida albicans and Candida parapsilosis. International Journal of Molecular Sciences. 2020; 21(3):850. https://doi.org/10.3390/ijms21030850
Chicago/Turabian StyleDostál, Jiří, Jan Blaha, Romana Hadravová, Martin Hubálek, Olga Heidingsfeld, and Iva Pichová. 2020. "Cellular Localization of Carbonic Anhydrase Nce103p in Candida albicans and Candida parapsilosis" International Journal of Molecular Sciences 21, no. 3: 850. https://doi.org/10.3390/ijms21030850
APA StyleDostál, J., Blaha, J., Hadravová, R., Hubálek, M., Heidingsfeld, O., & Pichová, I. (2020). Cellular Localization of Carbonic Anhydrase Nce103p in Candida albicans and Candida parapsilosis. International Journal of Molecular Sciences, 21(3), 850. https://doi.org/10.3390/ijms21030850