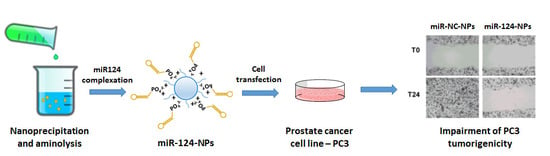
Cationic Polymer Nanoparticles-Mediated Delivery of miR-124 Impairs Tumorigenicity of Prostate Cancer Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Nanoparticles Synthesis and Characterization
2.2. Characterization of PHB-PEI NPs/miR-124 Complexes (miR-124 NPs)
2.3. Cellular Uptake of miR-124 NPs
2.4. Anti-Cancer Effects of miR-124 NPs
3. Materials and Methods
3.1. Materials
3.2. Synthesis and Functionalization of PHB NPs
3.3. Nanoparticles Characterization
3.4. PHB-PEI NPs Cytocompatibility
3.5. PHB-PEI NPs/miRNA Complex (miR-124 NPs) Formation and Characterization
3.6. Cell Culture
3.7. miRNAs Transient Transfection
3.8. RNA Isolation, Reverse Transcription, and Quantitative Real-Time PCR
3.9. Western Blotting
3.10. miR-124 NPs Cell Viability and Colony Forming Assay
3.11. Cellular Uptake of miRNA
3.12. miR-124 NPs Tumor Cell Migration and Invasion Assays
3.13. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saad, F.; Fizazi, K. Androgen Deprivation Therapy and Secondary Hormone Therapy in the Management of Hormone-sensitive and Castration-resistant Prostate Cancer. Urology 2015, 86, 852–861. [Google Scholar] [CrossRef] [PubMed]
- Nuhn, P.; De Bono, J.S.; Fizazi, K.; Freedland, S.J.; Grilli, M.; Kantoff, P.W.; Sonpavde, G.; Sternberg, C.N.; Yegnasubramanian, S.; Antonarakis, E.S. Update on Systemic Prostate Cancer Therapies: Management of Metastatic Castration-resistant Prostate Cancer in the Era of Precision Oncology. Eur. Urol. 2019, 75, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Vanacore, D.; Boccellino, M.; Rossetti, S.; Cavaliere, C.; D’Aniello, C.; Di Franco, R.; Romano, F.J.; Montanari, M.; La Mantia, E.; Piscitelli, R.; et al. Micrornas in prostate cancer: An overview. Oncotarget 2017, 8, 50240–50251. [Google Scholar] [CrossRef] [Green Version]
- Bryzgunova, O.E.; Konoshenko, M.Y.; Laktionov, P.P. MicroRNA-guided gene expression in prostate cancer: Literature and database overview. J. Gene Med. 2018, 20, e3016. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Bucci, J.; Chang, L.; Malouf, D.; Graham, P.; Li, Y. Targeting MicroRNAs in Prostate Cancer Radiotherapy. Theranostics 2017, 7, 3243–3259. [Google Scholar] [CrossRef] [Green Version]
- Kanwal, R.; Plaga, A.R.; Liu, X.; Shukla, G.C.; Gupta, S. MicroRNAs in prostate cancer: Functional role as biomarkers. Cancer Lett 2017, 407, 9–20. [Google Scholar] [CrossRef]
- Shukla, K.K.; Misra, S.; Pareek, P.; Mishra, V.; Singhal, B.; Sharma, P. Recent scenario of microRNA as diagnostic and prognostic biomarkers of prostate cancer. Urol. Oncol. 2017, 35, 92–101. [Google Scholar] [CrossRef]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef]
- Ganju, A.; Khan, S.; Hafeez, B.B.; Behrman, S.W.; Yallapu, M.M.; Chauhan, S.C.; Jaggi, M. miRNA nanotherapeutics for cancer. Drug Discov. Today 2017, 22, 424–432. [Google Scholar] [CrossRef] [Green Version]
- Mucaj, V.; Lee, S.S.; Skuli, N.; Giannoukos, D.N.; Qiu, B.; Eisinger-Mathason, T.S.K.; Nakazawa, M.S.; Shay, J.E.S.; Gopal, P.P.; Venneti, S.; et al. MicroRNA-124 expression counteracts pro-survival stress responses in glioblastoma. Oncogene 2015, 34, 2204–2214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Kim, H.J.; Park, C.K.; Kim, Y.-G.; Lee, H.-J.; Kim, J.-Y.; Kim, H.-H. MicroRNA-124 regulates osteoclast differentiation. Bone 2013, 56, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Luo, J.; Wang, B.; Wang, D.; Xie, X.; Yuan, L.; Guo, J.; Xi, S.; Gao, J.; Lin, X.; et al. Microrna-124 targets flotillin-1 to regulate proliferation and migration in breast cancer. Mol. Cancer 2013, 12, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Wang, S.; Zhang, W.; Qiu, J.; Shan, Y.; Yang, D.; Shen, B. Screening key microRNAs for castration-resistant prostate cancer based on miRNA/mRNA functional synergistic network. Oncotarget 2015, 6, 43819–43830. [Google Scholar] [CrossRef]
- Chu, M.; Chang, Y.; Guo, Y.; Wang, N.; Cui, J.; Gao, W.-Q. Regulation and methylation of tumor suppressor miR-124 by androgen receptor in prostate cancer cells. PLoS ONE 2015, 10, e0116197. [Google Scholar] [CrossRef]
- Shi, X.-B.; Ma, A.-H.; Xue, L.; Li, M.; Nguyen, H.G.; Yang, J.C.; Tepper, C.G.; Gandour-Edwards, R.; Evans, C.P.; Kung, H.-J.; et al. miR-124 and Androgen Receptor Signaling Inhibitors Repress Prostate Cancer Growth by Downregulating Androgen Receptor Splice Variants, EZH2, and Src. Cancer Res. 2015, 75, 5309–5317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.B.; Xue, L.; Ma, A.H.; Tepper, C.G.; Gandour-Edwards, R.; Kung, H.J.; de Vere White, R.W. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene 2013, 32, 4130–4138. [Google Scholar] [CrossRef] [Green Version]
- Valentino, A.; Calarco, A.; Di Salle, A.; Finicelli, M.; Crispi, S.; Calogero, R.A.; Riccardo, F.; Sciarra, A.; Gentilucci, A.; Galderisi, U.; et al. Deregulation of MicroRNAs mediated control of carnitine cycle in prostate cancer: Molecular basis and pathophysiological consequences. Oncogene 2017, 36, 6030–6040. [Google Scholar] [CrossRef]
- Hu, J.; Sheng, Y.; Shi, J.; Yu, B.; Yu, Z.; Liao, G. Long Circulating Polymeric Nanoparticles for Gene/Drug Delivery. Curr. Drug Metab. 2018, 19, 723–738. [Google Scholar] [CrossRef]
- Fujita, Y.; Kuwano, K.; Ochiya, T. Development of small RNA delivery systems for lung cancer therapy. Int. J. Mol. Sci. 2015, 16, 5254–5270. [Google Scholar] [CrossRef] [Green Version]
- Muthiah, M.; Park, I.-K.; Cho, C.-S. Nanoparticle-mediated delivery of therapeutic genes: Focus on miRNA therapeutics. Expert Opin. Drug Deliv. 2013, 10, 1259–1273. [Google Scholar] [CrossRef] [PubMed]
- Mauri, E.; Perale, G.; Rossi, F. Nanogel Functionalization: A Versatile Approach To Meet the Challenges of Drug and Gene Delivery. ACS Appl. Nano Mater. 2018, 1, 6525–6541. [Google Scholar] [CrossRef]
- Saraiva, C.; Paiva, J.; Santos, T.; Ferreira, L.; Bernardino, L. MicroRNA-124 loaded nanoparticles enhance brain repair in Parkinson’s disease. J. Control. Release 2016, 235, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Louw, A.M.; Kolar, M.K.; Novikova, L.N.; Kingham, P.J.; Wiberg, M.; Kjems, J.; Novikov, L.N. Chitosan polyplex mediated delivery of miRNA-124 reduces activation of microglial cells in vitro and in rat models of spinal cord injury. Nanomedicine 2016, 12, 643–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulze, J.; Kuhn, S.; Hendrikx, S.; Schulz-Siegmund, M.; Polte, T.; Aigner, A. Spray-Dried Nanoparticle-in-Microparticle Delivery Systems (NiMDS) for Gene Delivery, Comprising Polyethylenimine (PEI)-Based Nanoparticles in a Poly(Vinyl Alcohol) Matrix. Small 2018, 14. [Google Scholar] [CrossRef]
- Pandey, A.P.; Sawant, K.K. Polyethylenimine: A versatile, multifunctional non-viral vector for nucleic acid delivery. Mater. Sci. Eng. C 2016, 68, 904–918. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Wagner, E. Polymeric Carriers for Nucleic Acid Delivery: Current Designs and Future Directions. Biomacromolecules 2019, 20, 3613–3626. [Google Scholar] [CrossRef] [PubMed]
- Shrivastav, A.; Kim, H.-Y.; Kim, Y.-R. Advances in the applications of polyhydroxyalkanoate nanoparticles for novel drug delivery system. Biomed. Res. Int 2013, 2013, 581684. [Google Scholar] [CrossRef]
- Nigmatullin, R.; Thomas, P.; Lukasiewicz, B.; Puthussery, H.; Roy, I. Polyhydroxyalkanoates, a family of natural polymers, and their applications in drug delivery. J. Chem. Technol. Biotechnol. 2015, 90, 1209–1221. [Google Scholar] [CrossRef]
- Meng, D.-C.; Chen, G.-Q. Synthetic Biology of Polyhydroxyalkanoates (PHA). Adv. Biochem. Eng. Biotechnol. 2018, 162, 147–174. [Google Scholar] [CrossRef]
- Pişkin, E. Biodegradable polymers as biomaterials. J. Biomater. Sci. Polym. Ed. 1995, 6, 775–795. [Google Scholar] [CrossRef] [PubMed]
- Calarco, A.; Bosetti, M.; Margarucci, S.; Fusaro, L.; Nicolì, E.; Petillo, O.; Cannas, M.; Galderisi, U.; Peluso, G. The genotoxicity of PEI-based nanoparticles is reduced by acetylation of polyethylenimine amines in human primary cells. Toxicol. Lett. 2013, 218, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Höbel, S.; Aigner, A. Polyethylenimines for siRNA and miRNA delivery in vivo. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013, 5, 484–501. [Google Scholar] [CrossRef] [PubMed]
- D’Ayala, G.G.; Calarco, A.; Malinconico, M.; Laurienzo, P.; Petillo, O.; Torpedine, A.; Peluso, G. Cationic copolymers nanoparticles for nonviral gene vectors: Synthesis, characterization, and application in gene delivery. J. Biomed. Mater. Res. A 2010, 94, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Moret, I.; Esteban Peris, J.; Guillem, V.M.; Benet, M.; Revert, F.; Dasí, F.; Crespo, A.; Aliño, S.F. Stability of PEI-DNA and DOTAP-DNA complexes: Effect of alkaline pH, heparin and serum. J. Control. Release 2001, 76, 169–181. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, N. Cationic polymer optimization for efficient gene delivery. Mini Rev. Med. Chem. 2010, 10, 108–125. [Google Scholar] [CrossRef]
- Zhupanyn, P.; Ewe, A.; Büch, T.; Malek, A.; Rademacher, P.; Müller, C.; Reinert, A.; Jaimes, Y.; Aigner, A. Extracellular vesicle (ECV)-modified polyethylenimine (PEI) complexes for enhanced siRNA delivery in vitro and in vivo. J. Control. Release 2019, 319, 63–76. [Google Scholar] [CrossRef]
- Zhou, Y.; Yu, F.; Zhang, F.; Chen, G.; Wang, K.; Sun, M.; Li, J.; Oupický, D. Cyclam-Modified PEI for Combined VEGF siRNA Silencing and CXCR4 Inhibition To Treat Metastatic Breast Cancer. Biomacromolecules 2018, 19, 392–401. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, C. Tuning the Size of Poly(lactic-co-glycolic Acid) (PLGA) Nanoparticles Fabricated by Nanoprecipitation. Biotechnol. J. 2018, 13. [Google Scholar] [CrossRef]
- Croll, T.I.; O’Connor, A.J.; Stevens, G.W.; Cooper-White, J.J. Controllable surface modification of poly(lactic-co-glycolic acid) (PLGA) by hydrolysis or aminolysis I: Physical, chemical, and theoretical aspects. Biomacromolecules 2004, 5, 463–473. [Google Scholar] [CrossRef]
- Auriemma, M.; Piscitelli, A.; Pasquino, R.; Cerruti, P.; Malinconico, M.; Grizzuti, N. Blending poly(3-hydroxybutyrate) with tannic acid: Influence of a polyphenolic natural additive on the rheological and thermal behavior. Eur. Polym. J. 2015, 63, 123–131. [Google Scholar] [CrossRef]
- Lakard, B.; Herlem, G.; Lakard, S.; Antoniou, A.; Fahys, B. Urea potentiometric biosensor based on modified electrodes with urease immobilized on polyethylenimine films. Biosens. Bioelectron. 2004, 19, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- Godbey, W.T.; Wu, K.K.; Mikos, A.G. Poly(ethylenimine)-mediated gene delivery affects endothelial cell function and viability. Biomaterials 2001, 22, 471–480. [Google Scholar] [CrossRef]
- Kunath, K.; von Harpe, A.; Fischer, D.; Petersen, H.; Bickel, U.; Voigt, K.; Kissel, T. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: Comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J. Control. Release 2003, 89, 113–125. [Google Scholar] [CrossRef]
- Morachis, J.M.; Mahmoud, E.A.; Almutairi, A. Physical and chemical strategies for therapeutic delivery by using polymeric nanoparticles. Pharm. Rev. 2012, 64, 505–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murugan, K.; Choonara, Y.E.; Kumar, P.; Bijukumar, D.; du Toit, L.C.; Pillay, V. Parameters and characteristics governing cellular internalization and trans-barrier trafficking of nanostructures. Int. J. Nanomed. 2015, 10, 2191–2206. [Google Scholar] [CrossRef] [Green Version]
- Devulapally, R.; Sekar, N.M.; Sekar, T.V.; Foygel, K.; Massoud, T.F.; Willmann, J.K.; Paulmurugan, R. Polymer nanoparticles mediated codelivery of antimiR-10b and antimiR-21 for achieving triple negative breast cancer therapy. ACS Nano 2015, 9, 2290–2302. [Google Scholar] [CrossRef] [Green Version]
- Wong, L.-Y.; Xia, B.; Wolvetang, E.; Cooper-White, J. Targeted, Stimuli-Responsive Delivery of Plasmid DNA and miRNAs Using a Facile Self-Assembled Supramolecular Nanoparticle System. Biomacromolecules 2018, 19, 353–363. [Google Scholar] [CrossRef]
- Zadra, G.; Photopoulos, C.; Loda, M. The fat side of prostate cancer. Biochim. Biophys. Acta 2013, 1831, 1518–1532. [Google Scholar] [CrossRef] [Green Version]
- Long, J.; Zhang, C.-J.; Zhu, N.; Du, K.; Yin, Y.-F.; Tan, X.; Liao, D.-F.; Qin, L. Lipid metabolism and carcinogenesis, cancer development. Am. J. Cancer Res. 2018, 8, 778–791. [Google Scholar]
- Galbraith, L.; Leung, H.Y.; Ahmad, I. Lipid pathway deregulation in advanced prostate cancer. Pharm. Res. 2018, 131, 177–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, Q.; Zeng, F.; Liu, X.; Wang, Q.J.; Deng, F. Fatty acid oxidation and carnitine palmitoyltransferase I: Emerging therapeutic targets in cancer. Cell Death Dis. 2016, 7, e2226. [Google Scholar] [CrossRef] [PubMed]
- Melone, M.A.B.; Valentino, A.; Margarucci, S.; Galderisi, U.; Giordano, A.; Peluso, G. The carnitine system and cancer metabolic plasticity. Cell Death Dis. 2018, 9, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aiderus, A.; Black, M.A.; Dunbier, A.K. Fatty acid oxidation is associated with proliferation and prognosis in breast and other cancers. BMC Cancer 2018, 18, 805. [Google Scholar] [CrossRef] [PubMed]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoykova, G.E.; Schlaepfer, I.R. Lipid Metabolism and Endocrine Resistance in Prostate Cancer, and New Opportunities for Therapy. Int J. Mol. Sci. 2019, 20, 2626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlaepfer, I.R.; Rider, L.; Rodrigues, L.U.; Gijón, M.A.; Pac, C.T.; Romero, L.; Cimic, A.; Sirintrapun, S.J.; Glodé, L.M.; Eckel, R.H.; et al. Lipid catabolism via CPT1 as a therapeutic target for prostate cancer. Mol. Cancer 2014, 13, 2361–2371. [Google Scholar] [CrossRef] [Green Version]
- Ricciardi, M.R.; Mirabilii, S.; Allegretti, M.; Licchetta, R.; Calarco, A.; Torrisi, M.R.; Foà, R.; Nicolai, R.; Peluso, G.; Tafuri, A. Targeting the leukemia cell metabolism by the CPT1a inhibition: Functional preclinical effects in leukemias. Blood 2015, 126, 1925–1929. [Google Scholar] [CrossRef] [Green Version]
- Flaig, T.W.; Salzmann-Sullivan, M.; Su, L.-J.; Zhang, Z.; Joshi, M.; Gijón, M.A.; Kim, J.; Arcaroli, J.J.; Van Bokhoven, A.; Lucia, M.S.; et al. Lipid catabolism inhibition sensitizes prostate cancer cells to antiandrogen blockade. Oncotarget 2017, 8, 56051–56065. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-N.; Zeng, Z.-L.; Lu, J.; Wang, Y.; Liu, Z.-X.; He, M.-M.; Zhao, Q.; Wang, Z.-X.; Li, T.; Lu, Y.-X.; et al. CPT1A-mediated fatty acid oxidation promotes colorectal cancer cell metastasis by inhibiting anoikis. Oncogene 2018, 37, 6025–6040. [Google Scholar] [CrossRef]
- Fessi, H.; Puisieux, F.; Devissaguet, J.P.; Ammoury, N.; Benita, S. Nanocapsule formation by interfacial polymer deposition following solvent displacement. Int. J. Pharm. 1989, 55, R1–R4. [Google Scholar] [CrossRef]






| Genes | Forward primers (5′–3′) | Reverse primers (5′–3′) |
|---|---|---|
| ACTB | TTAGTTGCGTTACACCCTTTC | ACCTTCACCGTTCCAGTT |
| CPT1A | CTGGACAATACCTCGGAGCC | TCTAACGTCACGAAGAACGCT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conte, R.; Valentino, A.; Di Cristo, F.; Peluso, G.; Cerruti, P.; Di Salle, A.; Calarco, A. Cationic Polymer Nanoparticles-Mediated Delivery of miR-124 Impairs Tumorigenicity of Prostate Cancer Cells. Int. J. Mol. Sci. 2020, 21, 869. https://doi.org/10.3390/ijms21030869
Conte R, Valentino A, Di Cristo F, Peluso G, Cerruti P, Di Salle A, Calarco A. Cationic Polymer Nanoparticles-Mediated Delivery of miR-124 Impairs Tumorigenicity of Prostate Cancer Cells. International Journal of Molecular Sciences. 2020; 21(3):869. https://doi.org/10.3390/ijms21030869
Chicago/Turabian StyleConte, Raffaele, Anna Valentino, Francesca Di Cristo, Gianfranco Peluso, Pierfrancesco Cerruti, Anna Di Salle, and Anna Calarco. 2020. "Cationic Polymer Nanoparticles-Mediated Delivery of miR-124 Impairs Tumorigenicity of Prostate Cancer Cells" International Journal of Molecular Sciences 21, no. 3: 869. https://doi.org/10.3390/ijms21030869
APA StyleConte, R., Valentino, A., Di Cristo, F., Peluso, G., Cerruti, P., Di Salle, A., & Calarco, A. (2020). Cationic Polymer Nanoparticles-Mediated Delivery of miR-124 Impairs Tumorigenicity of Prostate Cancer Cells. International Journal of Molecular Sciences, 21(3), 869. https://doi.org/10.3390/ijms21030869