Influence of Salt Stress on Growth of Spermosphere Bacterial Communities in Different Peanut (Arachis hypogaea L.) Cultivars
Abstract
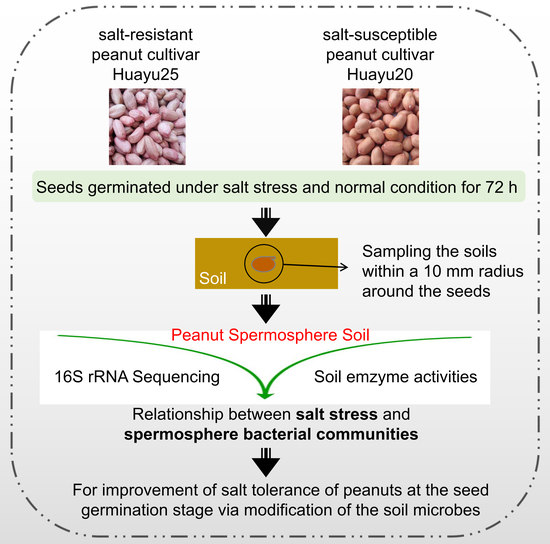
:1. Introduction
2. Results
2.1. Overall and Alpha Diversity Analysis of 16S rRNA Gene Sequencing Data
2.2. Differences in the Peanut Spermosphere Bacterial Community Structure
2.3. Beta Diversity Analysis of Bacterial Community
2.4. Specific Phylotypes of Peanut Spermosphere Modulate by Salt Stress
2.5. Metabolic Functional Features of the Peanut Spermosphere Bacterial Community
2.6. The Response of Peanut Spermosphere Soil Extracellular Enzyme Activities to Salt Stress
2.7. Quantitative PCR of Specific Bacterial Groups
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Seed Treatment
4.2. Samples of Spermosphere Compartments Collection and DNA Extraction
4.3. 16S rRNA Gene Sequencing
4.4. Bioinformatics Analysis
4.5. Alpha and Beta Diversity Analysis
4.6. LEfSe and Metabolic Functional Prediction
4.7. Quantification of Predominant Phyla or Genera in the Peanut Spermosphere Samples
4.8. Soil Enzyme Assays
4.9. Data Availability
4.10. Statistical Tests
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Huayu25 | salt-resistant peanut cultivar |
| SS25 | salt-treated spermosphere around Huayu25 |
| LDA | linear discriminant analysis |
| LEfSe | LDA effect size |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| qRT-PCR | quantitative real-time polymerase chain reaction |
References
- Dai, L.; Zhang, G.; Yu, Z.; Ding, H.; Xu, Y.; Zhang, Z. Effect of Drought Stress and Developmental Stages on Microbial Community Structure and Diversity in Peanut Rhizosphere Soil. Int. J. Mol. Sci. 2019, 20, 2265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Q.; Yu, Z.; Liu, P.; Zheng, H.; Xu, Y.; Sai, S.; Wu, Y.; Zheng, C. High Efficient Expression and Purification of Human Epidermal Growth Factor in Arachis hypogaea L. Int. J. Mol. Sci. 2019, 20, 2045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, L.; Wu, Y.; Huang, J.; Dai, X.; Lei, Y.; Yan, L.; Jiang, H.; Zhang, J.; Varshney, R.K.; Liao, B. Identification of ERF genes in peanuts and functional analysis of AhERF008 and AhERF019 in abiotic stress response. Funct. Integr. Genom. 2014, 14, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Bhaduri, D.; Meena, H.N.; Kalariya, K. External potassium (K+) application improves salinity tolerance by promoting Na+-exclusion, K+-accumulation and osmotic adjustment in contrasting peanut cultivars. Plant Physiol. Biochem. 2016, 103, 143–153. [Google Scholar] [CrossRef]
- Ibrahim, E. Seed priming to alleviate salinity stress in germinating seeds. J. Plant Physiol. 2016, 192, 38–46. [Google Scholar] [CrossRef]
- Zhang, C.; Luo, W.; Li, Y.; Zhang, X.; Bai, X.; Niu, Z.; Zhang, X.; Li, Z.; Wan, D. Transcriptomic Analysis of Seed Germination Under Salt Stress in Two Desert Sister Species (Populus euphratica and P. pruinosa). Front. Genet. 2019, 10, 231. [Google Scholar] [CrossRef]
- Xu, Y.; Yu, Z.; Zhang, S.; Wu, C.; Yang, G.; Yan, K.; Zheng, C.; Huang, J. CYSTM3 negatively regulates salt stress tolerance in Arabidopsis. Plant Mol. Boil. 2019, 99, 395–406. [Google Scholar] [CrossRef]
- Xu, Y.; Yu, Z.; Zhang, D.; Huang, J.; Wu, C.; Yang, G.; Yan, K.; Zhang, S.; Zheng, C. CYSTM, a Novel Non-Secreted Cysteine-Rich Peptide Family, Involved in Environmental Stresses in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 423–438. [Google Scholar] [CrossRef] [Green Version]
- Savvides, A.; Ali, S.; Tester, M.; Fotopoulos, V. Chemical Priming of Plants Against Multiple Abiotic Stresses: Mission Possible? Trends Plant Sci. 2016, 21, 329–340. [Google Scholar] [CrossRef] [Green Version]
- Bewley, J.D. Seed Germination and Dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Yang, P. Studies on the molecular mechanisms of seed germination. Proteomics 2015, 15, 1671–1679. [Google Scholar] [CrossRef] [PubMed]
- Holdsworth, M.J.; Bentsink, L.; Soppe, W.J.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuan, P.A.; Kumar, R.; Rehal, P.K.; Toora, P.K.; Ayele, B.T. Molecular Mechanisms Underlying Abscisic Acid/Gibberellin Balance in the Control of Seed Dormancy and Germination in Cereals. Front. Plant Sci. 2018, 9, 668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koornneef, M.; Jorna, M.L.; Brinkhorst-van der Swan, D.L.; Karssen, C.M. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 1982, 61, 385–393. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, D.; Xu, Y.; Jin, S.; Zhang, L.; Zhang, S.; Yang, G.; Huang, J.; Yan, K.; Wu, C.; et al. CEPR2 phosphorylates and accelerates the degradation of PYR/PYLs in Arabidopsis. J. Exp. Bot. 2019, 70, 5457–5469. [Google Scholar] [CrossRef] [Green Version]
- Léon-Kloosterziel, K.M.; Gil, M.A.; Ruijs, G.J.; Jacobsen, S.E.; E Olszewski, N.; Schwartz, S.H.; Zeevaart, J.A.D.; Koornneef, M. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996, 10, 655–661. [Google Scholar]
- Liu, Y.; Zuo, S.; Zou, Y.; Wang, J.; Song, W. Investigation on diversity and population succession dynamics of indigenous bacteria of the maize spermosphere. World J. Microbiol. Biotechnol. 2011, 28, 391–396. [Google Scholar] [CrossRef]
- Lopez-Velasco, G.; Carder, P.A.; Welbaum, G.; Ponder, M.A. Diversity of the spinach (Spinacia oleracea) spermosphere and phyllosphere bacterial communities. FEMS Microbiol. Lett. 2013, 346, 146–154. [Google Scholar] [CrossRef] [Green Version]
- Verona, O. The spermosphere. Ann. Inst. Pasteur (Paris) 1958, 95, 795–798. [Google Scholar]
- Nelson, E.B. Microbial dynamics and interactions in the spermosphere. Annu. Rev. Phytopathol. 2004, 42, 271–309. [Google Scholar] [CrossRef] [Green Version]
- Schiltz, S.; Gaillard, I.; Pawlicki-Jullian, N.; Thiombiano, B.; Mesnard, F.; Gontier, E. A review: What is the spermosphere and how can it be studied? J. Appl. Microbiol. 2015, 119, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, K.; Nelson, E.B. Differential Inactivation of Seed Exudate Stimulation of Pythium ultimum Sporangium Germination by Enterobacter cloacae Influences Biological Control Efficacy on Different Plant Species. Appl. Environ. Microbiol. 2003, 69, 1114–1120. [Google Scholar] [CrossRef] [Green Version]
- Windstam, S.; Nelson, E.B. Differential Interference with Pythium ultimum Sporangial Activation and Germination by Enterobacter cloacae in the Corn and Cucumber Spermospheres. Appl. Environ. Microbiol. 2008, 74, 4285–4291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.-T.; Li, Y.-N.; Wang, C.-Y.; Kim, K.S.; Wang, T.; Liu, S. Characteristics of the rhizosphere bacterial community across different cultivation years in saline–alkaline paddy soils of Songnen Plain of China. Can. J. Microbiol. 2018, 64, 925–936. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, R.R.; Pineda, R.P.; Barney, J.N.; Nilsen, E.T.; Barrett, J.E.; Williams, M.A. Plant Invasions Associated with Change in Root-Zone Microbial Community Structure and Diversity. PLoS ONE 2015, 10, e0141424. [Google Scholar] [CrossRef] [Green Version]
- Dindar, E.; Şağban, F.O.T.; Başkaya, H.S. Evaluation-of soil enzyme activities as soil quality indicators in sludge-amended soils. J. Environ. Boil. 2015, 36, 919–926. [Google Scholar]
- Yu, H.; Si, P.; Shao, W.; Qiao, X.; Yang, X.; Gao, D.; Wang, Z. Response of enzyme activities and microbial communities to soil amendment with sugar alcohols. Microbiology 2016, 5, 604–615. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.; Wang, X.; Yan, J.; Luo, L. Characterizing the Intra-Vineyard Variation of Soil Bacterial and Fungal Communities. Front. Microbiol. 2019, 10, 1239. [Google Scholar] [CrossRef] [Green Version]
- Yadav, B.K.; Tarafdar, J.C. Ability of Emericella rugulosa to mobilize unavailable P compounds during Pearl millet [Pennisetum glaucum (L.) R. Br.] crop under arid condition. Indian J. Microbiol. 2007, 47, 57–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-López, A.; Rogel, M.A.; Ormeño-Orrillo, E.; Martinez-Romero, J.C.; Martinez-Romero, E. Phaseolus vulgaris seed-borne endophytic community with novel bacterial species such as Rhizobium endophyticum sp. nov. Syst. Appl. Microbiol. 2010, 33, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Cankar, K.; Kraigher, H.; Ravnikar, M.; Rupnik, M. Bacterial endophytes from seeds of Norway spruce (Picea abiesL. Karst). FEMS Microbiol. Lett. 2005, 244, 341–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Wang, N.; Guo, X.; Zhang, Y.; Ye, B. Comparative analysis of bacterial community structure in the rhizosphere of maize by high-throughput pyrosequencing. PLoS ONE 2017, 12, e0178425. [Google Scholar] [CrossRef] [PubMed]
- Berge, O.; Guinebretière, M.-H.; Achouak, W.; Normand, P.; Heulin, T. Paenibacillus graminis sp. nov. and Paenibacillus odorifer sp. nov., isolated from plant roots, soil and food. Int. J. Syst. Evol. Microbiol. 2002, 52, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Elo, S.; Suominen, I.; Kämpfer, P.; Juhanoja, J.; Salkinoja-Salonen, M.; Haahtela, K. Paenibacillus borealis sp. nov., a nitrogen-fixing species isolated from spruce forest humus in Finland. Int. J. Syst. Evol. Microbiol. 2001, 51, 535–545. [Google Scholar] [CrossRef] [Green Version]
- Adesemoye, A.; Obini, M.; Ugoji, E. Comparison of plant growth-promotion with Pseudomonas aeruginosa and Bacillus subtilis in three vegetables. Braz. J. Microbiol. 2008, 39, 423–426. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Kim, N.-U.; Park, S.; Yoon, J.-H.; Ka, J.-O. Massilia chloroacetimidivorans sp. nov., a chloroacetamide herbicide-degrading bacterium isolated from soil. Antonie van Leeuwenhoek 2017, 63, 1297–1758. [Google Scholar] [CrossRef]
- Rodrıguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Singh, B.; Satyanarayana, T. Microbial phytases in phosphorus acquisition and plant growth promotion. Physiol. Mol. Boil. Plants 2011, 17, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Illarionova, E.S. Organic phosphorus of the soil and its mineralization. Boil. Bull. Acad. Sci. USSR 1978, 5, 293–299. [Google Scholar]
- Idriss, E.E.; Makarewicz, O.; Farouk, A.; Rosner, K.; Greiner, R.; Bochow, H.; Richter, T.; Borriss, R. Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth-promoting effect. Microbiology 2002, 148, 2097–2109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Ben Fekih, I.; Ma, Y.; Herzberg, M.; Zhang, C.; Li, Y.P.; Mazhar, S.H.; Bello, S.K.; Yang, N.; Su, J.; Xu, J.; et al. Draft Genome Sequence of Pseudarthrobacter sp. Strain AG30, Isolated from a Gold and Copper Mine in China. Microbiol. Resour. Announc. 2018, 7, e01329-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finger, S.; Godoy, F.; Wittwer, G.; Aranda, C.P.; Calderón, R.; Miranda, C.D. Purification and characterization of indochrome type blue pigment produced by Pseudarthrobacter sp. 34LCH1 isolated from Atacama desert. J. Ind. Microbiol. Biotechnol. 2019, 46, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Xie, B.; Wang, J.; He, W.; Wang, X.; Wei, G. County-Scale Spatial Distribution of Soil Enzyme Activities and Enzyme Activity Indices in Agricultural Land: Implications for Soil Quality Assessment. Sci. World J. 2014, 2014, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Zhang, S.; Han, P.; Hu, X.; Xie, L.; Li, Y.; Brooks, M.; Liao, X.; Qin, L. Soil Enzyme Activity in Soils Subjected to Flooding and the Effect on Nitrogen and Phosphorus Uptake by Oilseed Rape. Front. Plant Sci. 2019, 10, 368. [Google Scholar] [CrossRef] [Green Version]
- Tao, P.; Li, H.; Yu, Y.; Gu, J.; Lei, J. Ectoine and 5-hydroxyectoine accumulation in the halophile Virgibacillus halodenitrificans PDB-F2 in response to salt stress. Appl. Microbiol. Biotechnol. 2016, 100, 6779–6789. [Google Scholar] [CrossRef]
- Poplinger, M.; Shumilin, I.; Harries, D. Impact of trehalose on the activity of sodium and potassium chloride in aqueous solutions: Why trehalose is worth its salt. Food Chem. 2017, 237, 1209–1215. [Google Scholar] [CrossRef]
- Bharti, N.; Yadav, D.; Barnawal, D.; Maji, D.; Kalra, A. Exiguobacterium oxidotolerans, a halotolerant plant growth promoting rhizobacteria, improves yield and content of secondary metabolites in Bacopa monnieri (L.) Pennell under primary and secondary salt stress. World J. Microbiol. Biotechnol. 2012, 29, 379–387. [Google Scholar] [CrossRef]
- Ramakrishna, A.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar] [PubMed]
- Ilangumaran, G.; Smith, D.L. Plant Growth Promoting Rhizobacteria in Amelioration of Salinity Stress: A Systems Biology Perspective. Front. Plant Sci. 2017, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Weyens, N.; Beckers, B.; Schellingen, K.; Ceulemans, R.; Van Der Lelie, N.; Newman, L.; Taghavi, S.; Carleer, R.; Vangronsveld, J. The Potential of the Ni-Resistant TCE-Degrading Pseudomonas putida W619-TCE to Reduce Phytotoxicity and Improve Phytoremediation Efficiency of Poplar Cuttings on A Ni-TCE Co-Contamination. Int. J. Phytoremediation 2014, 17, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Geng, L.; Shao, G.-X.; Raymond, B.; Wang, M.-L.; Sun, X.-X.; Shu, C.-L.; Zhang, J. Subterranean infestation by Holotrichia parallela larvae is associated with changes in the peanut (Arachis hypogaea L.) rhizosphere microbiome. Microbiol. Res. 2018, 211, 13–20. [Google Scholar] [CrossRef]
- Evelin, H.; Kapoor, R.; Giri, B. Arbuscular mycorrhizal fungi in alleviation of salt stress: A review. Ann. Bot. 2009, 104, 1263–1280. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Kloepper, J.W.; Ryu, C.-M. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2009, 14, 1–4. [Google Scholar] [CrossRef]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; I Gordon, J.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Blaxter, M.; Mann, J.; Chapman, T.; Thomas, F.; Whitton, C.; Floyd, R.; Abebe, E. Defining operational taxonomic units using DNA barcode data. Philos. Trans. R. Soc. B Boil. Sci. 2005, 360, 1935–1943. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Huo, D.; Li, W.; Hu, Q.; Xu, C.; Liu, S.; Li, C. Metagenomic approach reveals microbial diversity and predictive microbial metabolic pathways in Yucha, a traditional Li fermented food. Sci. Rep. 2016, 6, 32524. [Google Scholar] [CrossRef]
- Zuo, Y.; Xie, W.; Pang, Y.; Li, T.; Li, Q.; Li, Y. Bacterial community composition in the gut content of Lampetra japonica revealed by 16S rRNA gene pyrosequencing. PLoS ONE 2017, 12, e0188919. [Google Scholar] [CrossRef]
- Chen, B.; Teh, B.-S.; Sun, C.; Hu, S.; Lu, X.; Boland, W.; Shao, Y. Biodiversity and Activity of the Gut Microbiota across the Life History of the Insect Herbivore Spodoptera littoralis. Sci. Rep. 2016, 6, 29505. [Google Scholar] [CrossRef] [PubMed]
- Maughan, H.; Wang, P.W.; Caballero, J.D.; Fung, P.; Gong, Y.; Donaldson, S.L.; Yuan, L.; Keshavjee, S.; Zhang, Y.; Yau, Y.C.W.; et al. Analysis of the Cystic Fibrosis Lung Microbiota via Serial Illumina Sequencing of Bacterial 16S rRNA Hypervariable Regions. PLoS ONE 2012, 7, e45791. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.T.; Clemente, J.C.; E Flores, G.; Walters, W.A.; Parfrey, L.W.; Knight, R.; Fierer, N. Global biogeography of highly diverse protistan communities in soil. ISME J. 2012, 7, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sheng, H.-F.; He, Y.; Wu, J.-Y.; Jiang, Y.-X.; Tam, N.; Zhou, H.-W. Comparison of the Levels of Bacterial Diversity in Freshwater, Intertidal Wetland, and Marine Sediments by Using Millions of Illumina Tags. Appl. Environ. Microbiol. 2012, 78, 8264–8271. [Google Scholar] [CrossRef] [Green Version]
- Jin, S.; Zhao, D.; Cai, C.; Song, D.; Shen, J.; Xu, A.; Qiao, Y.; Ran, Z.; Zheng, Q. Low-dose penicillin exposure in early life decreases Th17 and the susceptibility to DSS colitis in mice through gut microbiota modification. Sci. Rep. 2017, 7, 43662. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Long, M.; Li, H.; Gatesoupe, F.-J.; Zhang, X.; Zhang, Q.; Feng, D.; Li, A. Multi-Omics Analysis Reveals a Correlation between the Host Phylogeny, Gut Microbiota and Metabolite Profiles in Cyprinid Fishes. Front. Microbiol. 2017, 8, 454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Boil. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langille, M.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Li, Q.; Chen, J.; Wu, L.; Luo, X.; Li, N.; Arafat, Y.; Lin, S.; Lin, W. Belowground Interactions Impact the Soil Bacterial Community, Soil Fertility, and Crop Yield in Maize/Peanut Intercropping Systems. Int. J. Mol. Sci. 2018, 19, 622. [Google Scholar] [CrossRef] [PubMed] [Green Version]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Zhang, D.; Dai, L.; Ding, H.; Ci, D.; Qin, F.; Zhang, G.; Zhang, Z. Influence of Salt Stress on Growth of Spermosphere Bacterial Communities in Different Peanut (Arachis hypogaea L.) Cultivars. Int. J. Mol. Sci. 2020, 21, 2131. https://doi.org/10.3390/ijms21062131
Xu Y, Zhang D, Dai L, Ding H, Ci D, Qin F, Zhang G, Zhang Z. Influence of Salt Stress on Growth of Spermosphere Bacterial Communities in Different Peanut (Arachis hypogaea L.) Cultivars. International Journal of Molecular Sciences. 2020; 21(6):2131. https://doi.org/10.3390/ijms21062131
Chicago/Turabian StyleXu, Yang, Dai Zhang, Liangxiang Dai, Hong Ding, Dunwei Ci, Feifei Qin, Guanchu Zhang, and Zhimeng Zhang. 2020. "Influence of Salt Stress on Growth of Spermosphere Bacterial Communities in Different Peanut (Arachis hypogaea L.) Cultivars" International Journal of Molecular Sciences 21, no. 6: 2131. https://doi.org/10.3390/ijms21062131
APA StyleXu, Y., Zhang, D., Dai, L., Ding, H., Ci, D., Qin, F., Zhang, G., & Zhang, Z. (2020). Influence of Salt Stress on Growth of Spermosphere Bacterial Communities in Different Peanut (Arachis hypogaea L.) Cultivars. International Journal of Molecular Sciences, 21(6), 2131. https://doi.org/10.3390/ijms21062131