MicroRNAs and Extracellular Vesicles as Distinctive Biomarkers of Precocious and Advanced Stages of Breast Cancer Brain Metastases Development
Abstract
:1. Introduction
2. Results
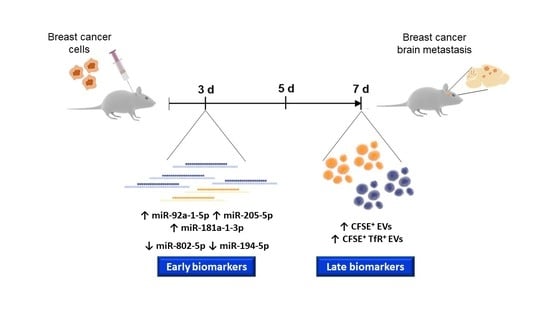
2.1. Circulating Brain Endothelial EVs Increase in Advanced Stages of BCBM
2.2. Circulating miRNAs Alterations Are Related with Their Brain Parenchyma Deregulation
2.3. miRNAs Are Differentially Released by BCCs, BMECs and During BCCs–BMECs Interaction
2.4. miR-194-5p and miR-205-5p Release Reflect Their Cellular Expression
3. Discussion
4. Materials and Methods
4.1. Mouse Model of BCBM
4.2. Cell Model of BCBM
4.3. Collection of Plasma
4.4. Collection of Brains
4.5. Nanoparticle Tracking
4.6. EVs Staining and Preparation
4.7. Flow Cytometry
4.8. RT-qPCR
4.9. In Situ Hybridization (ISH)
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 68, 1–41. [Google Scholar] [CrossRef]
- Branco, V.; Pimentel, J.; Brito, M.A.; Carvalho, C. Thioredoxin, Glutathione and Related Molecules in Tumors of the Nervous System. Curr. Med. Chem. 2020, 27, 1878–1900. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, E.M.; Anders, C.K. Understanding patterns of brain metastasis in breast cancer and designing rational therapeutic strategies. Ann. Transl. Med. 2018, 6, 163. [Google Scholar] [CrossRef] [PubMed]
- Custódio-Santos, T.; Videira, M.; Brito, M.A. Brain metastasization of breast cancer. BBA Rev. Cancer 2017, 1868, 132–147. [Google Scholar] [CrossRef] [PubMed]
- Sauter, E.R. Reliable Biomarkers to Identify New and Recurrent Cancer. Eur. J. Breast Health 2017, 13, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Ferracin, M.; Lupini, L.; Salamon, I.; Saccenti, E.; Zanzi, M.V.; Rocchi, A.; Da Ros, L.; Zagatti, B.; Musa, G.; Bassi, C.; et al. Absolute quantification of cell-free microRNAs in cancer patients. Oncotarget 2015, 6, 14545–14555. [Google Scholar] [CrossRef] [Green Version]
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun. Biol. 2019, 2, 325. [Google Scholar] [CrossRef] [Green Version]
- Sereno, M.; Videira, M.; Wilhelm, I.; Krizbai, I.A.; Brito, M.A. miRNAs in Health and Disease: A Focus on the Breast Cancer Metastatic Cascade towards the Brain. Cells 2020, 9, 1790. [Google Scholar] [CrossRef]
- Tenreiro, M.M.; Ferreira, R.; Bernardino, L.; Brito, M.A. Cellular response of the blood-brain barrier to injury: Potential biomarkers and therapeutic targets for brain regeneration. Neurobiol. Dis. 2016, 91, 262–273. [Google Scholar] [CrossRef]
- Couto, N.; Caja, S.; Maia, J.; Strano Moraes, M.C.; Costa-Silva, B. Exosomes as emerging players in cancer biology. Biochimie 2018, 155, 2–10. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Möller, A.; Lobb, R.J. The evolving translational potential of small extracellular vesicles in cancer. Nat. Rev. Cancer 2020, 20, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Li, Z.; Zhou, K.; Feng, N. Exosomes as Carriers for Antitumor Therapy. ACS Biomater. Sci. Eng. 2019, 5, 4870–4881. [Google Scholar] [CrossRef] [PubMed]
- Sabatier, F.; Camoin-Jau, L.; Anfosso, F.; Sampol, J.; Dignat-George, F. Circulating endothelial cells, microparticles and progenitors: Key players towards the definition of vascular competence. J. Cell Mol. Med. 2009, 13, 454–471. [Google Scholar] [CrossRef]
- Ramirez, S.H.; Andrews, A.M.; Paul, D.; Pachter, J.S. Extracellular vesicles: Mediators and biomarkers of pathology along CNS barriers. Fluids Barriers CNS 2018, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.L.; Brites, D.; Brito, M.A. Looking at the blood-brain barrier: Molecular anatomy and possible investigation approaches. Brain Res. Rev. 2010, 64, 328–363. [Google Scholar] [CrossRef]
- Hromada, C.; Mühleder, S.; Grillari, J.; Redl, H.; Holnthoner, W. Endothelial Extracellular Vesicles—Promises and Challenges. Front. Physiol. 2017, 8, 275. [Google Scholar] [CrossRef]
- Haskó, J.; Fazakas, C.; Molnár, K.; Mészáros, Á.; Patai, R.; Szabó, G.; Erdélyi, F.; Nyúl-Tóth, Á.; Győri, F.; Kozma, M.; et al. Response of the neurovascular unit to brain metastatic breast cancer cells. Acta Neuropathol. Commun. 2019, 7, 133. [Google Scholar] [CrossRef]
- Patel, M.; De Paoli, S.H.; Elhelu, O.K.; Farooq, S.; Simak, J. Cell membrane disintegration and extracellular vesicle release in a model of different size and charge PAMAM dendrimers effects on cultured endothelial cells. Nanotoxicology 2019, 13, 664–681. [Google Scholar] [CrossRef]
- Patching, S.G. Glucose Transporters at the Blood-Brain Barrier: Function, Regulation and Gateways for Drug Delivery. Mol. Neurobiol. 2017, 54, 1046–1077. [Google Scholar] [CrossRef]
- Pardridge, W.M.; Boado, R.J.; Farrell, C.R. Brain-type glucose transporter (GLUT-1) is selectively localized to the blood-brain barrier. Studies with quantitative western blotting and in situ hybridization. J. Biol. Chem. 1990, 265, 18035–18040. [Google Scholar] [CrossRef]
- Jefferies, W.A.; Brandon, M.R.; Hunt, S.V.; Williams, A.F.; Gatter, K.C.; Mason, D.Y. Transferrin receptor on endothelium of brain capillaries. Nature 1984, 312, 162–163. [Google Scholar] [CrossRef] [PubMed]
- Leitner, D.F.; Connor, J.R. Functional roles of transferrin in the brain. Biochim. Biophys. Acta 2012, 1820, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Figueira, I.; Galego, S.; Custódio-Santos, T.; Vicente, R.; Molnár, K.; Haskó, J.; Malhó, R.; Videira, M.; Wilhelm, I.; Krizbai, I.; et al. Picturing Breast Cancer Brain Metastasis Development to Unravel Molecular Players and Cellular Crosstalk. Cancers 2021, 13, 910. [Google Scholar] [CrossRef] [PubMed]
- Jay, C.; Nemunaitis, J.; Chen, P.; Fulgham, P.; Tong, A.W. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007, 26, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, A.A.; Larcher, L.M.; Chen, S.; Veedu, R.N. Therapeutically Significant MicroRNAs in Primary and Metastatic Brain Malignancies. Cancers 2020, 12, 2534. [Google Scholar] [CrossRef] [PubMed]
- Loh, H.-Y.; Norman, B.P.; Lai, K.-S.; Rahman, N.M.A.N.A.; Alitheen, N.B.M.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [Green Version]
- Ono, S.; Lam, S.; Nagahara, M.; Hoon, D.S.B. Circulating microRNA Biomarkers as Liquid Biopsy for Cancer Patients: Pros and Cons of Current Assays. J. Clin. Med. 2015, 4, 1890–1907. [Google Scholar] [CrossRef] [Green Version]
- Sereno, M.; Haskó, J.; Molnár, K.; Medina, S.J.; Reisz, Z.; Malhó, R.; Videira, M.; Tiszlavicz, L.; Booth, S.A.; Wilhelm, I.; et al. Downregulation of circulating miR 802-5p and miR 194-5p and upregulation of brain MEF2C along breast cancer brain metastasization. Mol. Oncol. 2020, 14, 520–538. [Google Scholar] [CrossRef] [Green Version]
- Tao, K.; Fang, M.; Alroy, J.; Sahagian, G.G. Imagable 4T1 model for the study of late stage breast cancer. BMC Cancer 2008, 8, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maia, J.; Batista, S.; Couto, N.; Gregório, A.C.; Bodo, C.; Elzanowska, J.; Strano Moraes, M.C.; Costa-Silva, B. Employing Flow Cytometry to Extracellular Vesicles Sample Microvolume Analysis and Quality Control. Front. Cell Dev. Biol. 2020, 8, 593750. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, M.; Sugaya, K. DNA Associated with Circulating Exosomes as a Biomarker for Glioma. Genes 2020, 11, 1276. [Google Scholar] [CrossRef]
- Duffy, M.J.; Sturgeon, C.M.; Sölétormos, G.; Barak, V.; Molina, R.; Hayes, D.F.; Diamandis, E.P.; Bossuyt, P.M.M. Validation of New Cancer Biomarkers: A Position Statement from the European Group on Tumor Markers. Clin. Chem. 2015, 61, 809–820. [Google Scholar] [CrossRef] [Green Version]
- Galindo-Hernandez, O.; Villegas-Comonfort, S.; Candanedo, F.; González-Vázquez, M.-C.; Chavez-Ocaña, S.; Jimenez-Villanueva, X.; Sierra-Martinez, M.; Salazar, E.P. Elevated Concentration of Microvesicles Isolated from Peripheral Blood in Breast Cancer Patients. Arch. Med. Res. 2013, 44, 208–214. [Google Scholar] [CrossRef]
- Haqqani, A.S.; Delaney, C.E.; Tremblay, T.-L.; Sodja, C.; Sandhu, J.K.; Stanimirovic, D.B. Method for isolation and molecular characterization of extracellular microvesicles released from brain endothelial cells. Fluids Barriers CNS 2013, 10, 4. [Google Scholar] [CrossRef]
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef]
- Toth, B.; Nieuwland, R.; Liebhardt, S.; Ditsch, N.; Steinig, K.; Stieber, P.; Rank, A.; Göhring, P.; Thaler, C.J.; Friese, K.; et al. Circulating microparticles in breast cancer patients: A comparative analysis with established biomarkers. Anticancer Res. 2008, 28, 1107–1112. [Google Scholar]
- García Garre, E.; Luengo Gil, G.; Montoro García, S.; Gonzalez Billalabeitia, E.; Zafra Poves, M.; García Martinez, E.; Roldán Schilling, V.; Navarro Manzano, E.; Ivars Rubio, A.; Lip, G.Y.H.; et al. Circulating small-sized endothelial microparticles as predictors of clinical outcome after chemotherapy for breast cancer: An exploratory analysis. Breast Cancer Res. Treat. 2018, 169, 83–92. [Google Scholar] [CrossRef]
- Risha, Y.; Minic, Z.; Ghobadloo, S.M.; Berezovski, M.V. The proteomic analysis of breast cell line exosomes reveals disease patterns and potential biomarkers. Sci. Rep. 2020, 10, 13572. [Google Scholar] [CrossRef]
- Pragallapati, S.; Manyam, R. Glucose transporter 1 in health and disease. J. Oral Maxillofac. Pathol. 2019, 23, 443–449. [Google Scholar] [CrossRef]
- Wang, J.; Ye, C.; Chen, C.; Xiong, H.; Xie, B.; Zhou, J.; Chen, Y.; Zheng, S.; Wang, L. Glucose transporter GLUT1 expression and clinical outcome in solid tumors: A systematic review and meta-analysis. Oncotarget 2017, 8, 16875–16886. [Google Scholar] [CrossRef] [Green Version]
- Jiwa, L.S.; van Diest, P.J.; Hoefnagel, L.D.; Wesseling, J.; Wesseling, P.; Dutch Distant Breast Cancer Metastases, C.; Moelans, C.B. Upregulation of Claudin-4, CAIX and GLUT-1 in distant breast cancer metastases. BMC Cancer 2014, 14, 864. [Google Scholar] [CrossRef] [Green Version]
- Keel, S.B.; Doty, R.; Liu, L.; Nemeth, E.; Cherian, S.; Ganz, T.; Abkowitz, J.L. Evidence that the expression of transferrin receptor 1 on erythroid marrow cells mediates hepcidin suppression in the liver. Exp. Hematol. 2015, 43, 469–478.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habashy, H.O.; Powe, D.G.; Staka, C.M.; Rakha, E.A.; Ball, G.; Green, A.R.; Aleskandarany, M.; Paish, E.C.; Douglas Macmillan, R.; Nicholson, R.I.; et al. Transferrin receptor (CD71) is a marker of poor prognosis in breast cancer and can predict response to tamoxifen. Breast Cancer Res. Treat. 2009, 119, 283. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Batagov, A.O.; Schinelli, S.; Wang, J.; Wang, Y.; El Fatimy, R.; Rabinovsky, R.; Balaj, L.; Chen, C.C.; Hochberg, F.; et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017, 8, 1145. [Google Scholar] [CrossRef] [Green Version]
- Karimi, N.; Cvjetkovic, A.; Jang, S.C.; Crescitelli, R.; Hosseinpour Feizi, M.A.; Nieuwland, R.; Lötvall, J.; Lässer, C. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell. Mol. Life Sci. CMLS 2018, 75, 2873–2886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radojicic, J.; Zaravinos, A.; Vrekoussis, T.; Kafousi, M.; Spandidos, D.A.; Stathopoulos, E.N. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle 2011, 10, 507–517. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, J. MicroRNA-mediated breast cancer metastasis: From primary site to distant organs. Oncogene 2012, 31, 2499–2511. [Google Scholar] [CrossRef] [Green Version]
- Heneghan, H.M.; Miller, N.; Kelly, R.; Newell, J.; Kerin, M.J. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist 2010, 15, 673–682. [Google Scholar] [CrossRef] [Green Version]
- Ng, E.K.; Li, R.; Shin, V.Y.; Jin, H.C.; Leung, C.P.; Ma, E.S.; Pang, R.; Chua, D.; Chu, K.M.; Law, W.L.; et al. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS ONE 2013, 8, e53141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Zhou, Y.; Xia, T.; Zhou, X.; Huang, Z.; Zhang, H.; Zhu, W.; Ding, Q.; Wang, S. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res. Treat. 2018, 170, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.Y.; Gong, H.T.; Li, B.F.; Lv, C.L.; Wang, H.T.; Zhou, H.H.; Li, X.X.; Xie, S.Y.; Jiang, B.F. Higher expression of circulating miR-182 as a novel biomarker for breast cancer. Oncol. Lett. 2013, 6, 1681–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, H.; Gallagher, D.; Schmitt, S.; Pessetto, Z.Y.; Fan, F.; Godwin, A.K.; Tawfik, O. Role of miR-139 as a surrogate marker for tumor aggression in breast cancer. Human Pathol. 2017, 61, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, A.C.; Xu, Z.; Weinberg, C.R.; Getts, R.C.; Wade, P.A.; DeRoo, L.A.; Sandler, D.P.; Taylor, J.A. Serum microRNA expression as an early marker for breast cancer risk in prospectively collected samples from the Sister Study cohort. Breast Cancer Res. BCR 2013, 15, R42. [Google Scholar] [CrossRef] [Green Version]
- Yuan, F.; Wang, W. MicroRNA-802 suppresses breast cancer proliferation through downregulation of FoxM1. Mol. Med. Rep. 2015, 12, 4647–4651. [Google Scholar] [CrossRef] [Green Version]
- Si, H.; Sun, X.; Chen, Y.; Cao, Y.; Chen, S.; Wang, H.; Hu, C. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J. Cancer Res. Clin. Oncol. 2013, 139, 223–229. [Google Scholar] [CrossRef] [Green Version]
- Garcia, R.; Godinho-Pereira, J.; Figueira, I.; Malhó, R.; Brito, M.A. Replicating the blood-brain barrier properties in an in vitro model: Effects of hydrocortisone and/or shear stress. Arch. Anat. 2019, 8, 4–20. [Google Scholar]
- Guo, J.; Duan, Z.; Zhang, C.; Wang, W.; He, H.; Liu, Y.; Wu, P.; Wang, S.; Song, M.; Chen, H.; et al. Mouse 4T1 Breast Cancer Cell–Derived Exosomes Induce Proinflammatory Cytokine Production in Macrophages via miR-183. J. Immunol. 2020, 205, 2916–2925. [Google Scholar] [CrossRef]
- Song, C.; Liu, L.; Pei, X.; Liu, X.; Yang, L.; Ye, F.; Xie, X.; Chen, J.; Tang, H.; Xie, X. miR-200c inhibits breast cancer proliferation by targeting KRAS. Oncotarget 2015, 6, 34968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tominaga, N.; Kosaka, N.; Ono, M.; Katsuda, T.; Yoshioka, Y.; Tamura, K.; Lötvall, J.; Nakagama, H.; Ochiya, T. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat. Commun. 2015, 6, 6716. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin Andrew, R.; et al. Cancer-Secreted miR-105 Destroys Vascular Endothelial Barriers to Promote Metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [Green Version]
- Di Modica, M.; Regondi, V.; Sandri, M.; Iorio, M.V.; Zanetti, A.; Tagliabue, E.; Casalini, P.; Triulzi, T. Breast cancer-secreted miR-939 downregulates VE-cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett. 2017, 384, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Zhu, S.; Mo, Y.-Y. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009, 19, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Humphries, B.; Yang, C.; Wang, Z. MiR-205 Dysregulations in Breast Cancer: The Complexity and Opportunities. Noncoding RNA 2019, 5, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Yu, M.; Zhao, K.; He, M.; Ge, W.; Sun, Y.; Wang, Y.; Sun, H.; Hu, Y. Upregulation of MiR-205 under hypoxia promotes epithelial-mesenchymal transition by targeting ASPP2. Cell Death Dis. 2016, 7, e2517. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ren, S.Y.; Qu, Y.; Liu, C.; Zhang, Y.; Li, X.Q.; Ma, H. MiR-194-5p inhibited metastasis and EMT of nephroblastoma cells through targeting Crk. Kaohsiung J. Med. Sci. 2020, 36, 265–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; Xiao, Z.; Zhang, S. Knockdown of miR-194-5p inhibits cell proliferation, migration and invasion in breast cancer by regulating the Wnt/β-catenin signaling pathway. Int. J. Mol. Med. 2018, 42, 3355–3363. [Google Scholar] [CrossRef] [Green Version]
- Le, X.-F.; Almeida, M.I.; Mao, W.; Spizzo, R.; Rossi, S.; Nicoloso, M.S.; Zhang, S.; Wu, Y.; Calin, G.A.; Bast, R.C., Jr. Modulation of MicroRNA-194 and Cell Migration by HER2-Targeting Trastuzumab in Breast Cancer. PLoS ONE 2012, 7, e41170. [Google Scholar] [CrossRef] [Green Version]
- Mao, Q.; Lv, M.; Li, L.; Sun, Y.; Liu, S.; Shen, Y.; Liu, Z.; Luo, S. Long intergenic noncoding RNA 00641 inhibits breast cancer cell proliferation, migration, and invasion by sponging miR-194-5p. J. Cell Physiol. 2020, 235, 2668–2675. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.D.; Iyer, A.M.; van Scheppingen, J.; Bongaarts, A.; Anink, J.J.; Janssen, B.; Zimmer, T.S.; Spliet, W.G.; van Rijen, P.C.; Jansen, F.E.; et al. Coding and small non-coding transcriptional landscape of tuberous sclerosis complex cortical tubers: Implications for pathophysiology and treatment. Sci. Rep. 2017, 7, 8089. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Figueira, I.; Godinho-Pereira, J.; Galego, S.; Maia, J.; Haskó, J.; Molnár, K.; Malhó, R.; Costa-Silva, B.; Wilhelm, I.; Krizbai, I.A.; et al. MicroRNAs and Extracellular Vesicles as Distinctive Biomarkers of Precocious and Advanced Stages of Breast Cancer Brain Metastases Development. Int. J. Mol. Sci. 2021, 22, 5214. https://doi.org/10.3390/ijms22105214
Figueira I, Godinho-Pereira J, Galego S, Maia J, Haskó J, Molnár K, Malhó R, Costa-Silva B, Wilhelm I, Krizbai IA, et al. MicroRNAs and Extracellular Vesicles as Distinctive Biomarkers of Precocious and Advanced Stages of Breast Cancer Brain Metastases Development. International Journal of Molecular Sciences. 2021; 22(10):5214. https://doi.org/10.3390/ijms22105214
Chicago/Turabian StyleFigueira, Inês, Joana Godinho-Pereira, Sofia Galego, Joana Maia, János Haskó, Kinga Molnár, Rui Malhó, Bruno Costa-Silva, Imola Wilhelm, István A. Krizbai, and et al. 2021. "MicroRNAs and Extracellular Vesicles as Distinctive Biomarkers of Precocious and Advanced Stages of Breast Cancer Brain Metastases Development" International Journal of Molecular Sciences 22, no. 10: 5214. https://doi.org/10.3390/ijms22105214
APA StyleFigueira, I., Godinho-Pereira, J., Galego, S., Maia, J., Haskó, J., Molnár, K., Malhó, R., Costa-Silva, B., Wilhelm, I., Krizbai, I. A., & Brito, M. A. (2021). MicroRNAs and Extracellular Vesicles as Distinctive Biomarkers of Precocious and Advanced Stages of Breast Cancer Brain Metastases Development. International Journal of Molecular Sciences, 22(10), 5214. https://doi.org/10.3390/ijms22105214