Null cyp1b1 Activity in Zebrafish Leads to Variable Craniofacial Defects Associated with Altered Expression of Extracellular Matrix and Lipid Metabolism Genes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
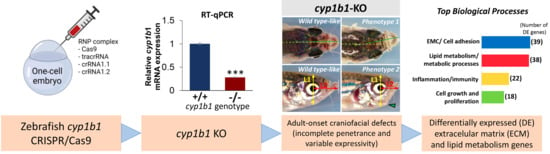
2.1. Generation and Characterization of a Cyp1b1-KO Zebrafish Line
2.2. F0 Embryo Development Delay and Variable Craniofacial Defects in Adult Zebrafish due to Cyp1b1 LoF
2.3. Comparison of Gene Expression Profiles of Cyp1b-KO and Wildtype Zebrafish
2.3.1. Functional Enrichment Analysis of DEGs
2.3.2. Validation of RNA-Seq Results
3. Discussion
4. Materials and Methods
4.1. Zebrafish Embryo Management
4.2. Cas9 Gene Editing
4.3. Genotyping
4.4. RT-qPCR
4.5. Fluorescent In Situ Hybridization
4.6. Examination of Mutant Phenotypes by Light Microscopy
4.7. RNA Preparation and Transcriptome Analysis
4.8. In Silico Analysis
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shimada, T.; Watanabe, J.; Inoue, K.; Guengerich, F.P.; Gillam, E.M. Specificity of 17beta-oestradiol and benzo[a]pyrene oxidation by polymorphic human cytochrome P4501B1 variants substituted at residues 48, 119 and 432. Xenobiotica 2001, 31, 163–176. [Google Scholar] [CrossRef]
- Choudhary, D.; Jansson, I.; Stoilov, I.; Sarfarazi, M.; Schenkman, J.B. Metabolism of retinoids and arachidonic acid by human and mouse cytochrome p450 1B1. Drug Metab. Dispos. 2004, 32, 840–847. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Howald, W.N.; Juchau, M.R. Biosynthesis of all-trans-retinoic acid from all-trans-retinol: Catalysis of all-trans-retinol oxidation by human P-450 cytochromes. Drug Metab. Dispos. 2000, 28, 315–322. [Google Scholar] [PubMed]
- Shimada, T.; Watanabe, J.; Kawajiri, K.; Sutter, T.R.; Guengerich, F.P.; Gillam, E.M.; Inoue, K. Catalytic properties of polymorphic human cytochrome P450 1B1 variants. Carcinogenesis 1999, 20, 1607–1614. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.M.; Wo, Y.-Y.P.; Stewart, J.; Hawkins, A.L.; Griffin, C.A.; Sutter, T.R.; Greenlee, W.F. Isolation and Characterization of the Human Cytochrome P450 CYP1B1 Gene. J. Biol. Chem. 1996, 271, 28324–28330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansal, S.; Leu, A.N.; Gonzalez, F.J.; Guengerich, F.P.; Chowdhury, A.R.; Anandatheerthavarada, H.K.; Avadhani, N.G. Mitochondrial Targeting of Cytochrome P450 (CYP) 1B1 and Its Role in Polycyclic Aromatic Hydrocarbon-induced Mitochondrial Dysfunction. J. Biol. Chem. 2014, 289, 9936–9951. [Google Scholar] [CrossRef] [Green Version]
- Achary, M.S.; Reddy, A.B.M.; Chakrabarti, S.; Panicker, S.G.; Mandal, A.K.; Ahmed, N.; Balasubramanian, R.; Hasnain, S.E.; Nagarajaram, H.A. Disease-Causing Mutations in Proteins: Structural Analysis of the CYP1B1 Mutations Causing Primary Congenital Glaucoma in Humans. Biophys. J. 2006, 91, 4329–4339. [Google Scholar] [CrossRef] [Green Version]
- Sarfarazi, M.; Akarsu, A.N.; Hossain, A.; Turacli, M.E.; Aktan, S.G.; Barsoum-Homsy, M.; Chevrette, L.; Sayli, B.S. Assignment of a locus (GLC3A) for primary congenital glaucoma (Buphthalmos) to 2p21 and evidence for genetic heterogeneity. Genomics 1995, 30, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Bejjani, B.A.; Lewis, R.A.; Tomey, K.F.; Anderson, K.L.; Dueker, D.K.; Jabak, M.; Astle, W.F.; Otterud, B.; Leppert, M.; Lupski, J.R. Mutations in CYP1B1, the Gene for Cytochrome P4501B1, Are the Predominant Cause of Primary Congenital Glaucoma in Saudi Arabia. Am. J. Hum. Genet. 1998, 62, 325–333. [Google Scholar] [CrossRef] [Green Version]
- Mashima, Y.; Suzuki, Y.; Sergeev, Y.; Ohtake, Y.; Tanino, T.; Kimura, I.; Miyata, H.; Aihara, M.; Tanihara, H.; Inatani, M.; et al. Novel cytochrome P4501B1 (CYP1B1) gene mutations in Japanese patients with primary congenital glaucoma. Investig. Ophthalmol. Vis. Sci. 2001, 42, 2211–2216. [Google Scholar]
- Panicker, S.G.; Reddy, A.B.M.; Mandal, A.K.; Ahmed, N.; Nagarajaram, H.A.; Hasnain, S.E.; Balasubramanian, R. Identification of novel mutations causing familial primary congenital glaucoma in Indian pedigrees. Investig. Ophthalmol. Vis. Sci. 2002, 43, 1358–1366. [Google Scholar]
- López-Garrido, M.-P.; Medina-Trillo, C.; Morales-Fernandez, L.; Garcia-Feijoo, J.; Martínez-De-La-Casa, J.-M.; García-Antón, M.; Escribano, J. Null CYP1B1 Genotypes in Primary Congenital and Nondominant Juvenile Glaucoma. Ophthalmology 2013, 120, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.L.; Walton, D.S. Primary Congenital Glaucoma: 2004 Update. J. Pediatr. Ophthalmol. Strabismus 2004, 41, 271–288. [Google Scholar] [CrossRef] [PubMed]
- Stoilov, I.; Akarsu, A.N.; Sarfarazi, M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum. Mol. Genet. 1997, 6, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Maul, E.; Strozzi, L.; Muñoz, C.; Reyes, C. The Outflow Pathway in Congenital Glaucoma. Am. J. Ophthalmol. 1980, 89, 667–675. [Google Scholar] [CrossRef]
- Anderson, D.R. The development of the trabecular meshwork and its abnormality in primary infantile glaucoma. Trans. Am. Ophthalmol. Soc. 1981, 79, 458–485. [Google Scholar]
- Kupfer, C.; Kaiser-Kupfer, M.I.; Kuwabara, T. Histopathology of abnormalities of the anterior chamber with glaucoma. Trans. Am. Ophthalmol. Soc. 1986, 84, 71–84. [Google Scholar]
- Antón, M.T.G.; Salazar, J.J.; De Hoz, R.; Rojas, B.; Ramírez, A.I.; Triviño, A.; Aroca-Aguilar, J.-D.; García-Feijoo, J.; Escribano, J.; Ramírez, J.M. Goniodysgenesis variability and activity of CYP1B1 genotypes in primary congenital glaucoma. PLoS ONE 2017, 12, e0176386. [Google Scholar] [CrossRef] [Green Version]
- Narooie-Nejad, M.; Paylakhi, S.H.; Shojaee, S.; Fazlali, Z.; Kanavi, M.R.; Nilforushan, N.; Yazdani, S.; Babrzadeh, F.; Suri, F.; Ronaghi, M.; et al. Loss of function mutations in the gene encoding latent transforming growth factor beta binding protein 2, LTBP2, cause primary congenital glaucoma. Hum. Mol. Genet. 2009, 18, 3969–3977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, M.; McKibbin, M.; Booth, A.; Parry, D.A.; Jain, P.; Riazuddin, S.A.; Hejtmancik, J.F.; Khan, S.N.; Firasat, S.; Shires, M.; et al. Null Mutations in LTBP2 Cause Primary Congenital Glaucoma. Am. J. Hum. Genet. 2009, 84, 664–671. [Google Scholar] [CrossRef] [Green Version]
- Kaur, K.; Reddy, A.; Mukhopadhyay, A.; Mandal, A.; Hasnain, S.; Ray, K.; Thomas, R.; Balasubramanian, D.; Chakrabarti, S. Myocilin gene implicated in primary congenital glaucoma. Clin. Genet. 2005, 67, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Souma, T.; Tompson, S.W.; Thomson, B.R.; Siggs, O.M.; Kizhatil, K.; Yamaguchi, S.; Feng, L.; Limviphuvadh, V.; Whisenhunt, K.N.; Maurer-Stroh, S.; et al. Angiopoietin receptor TEK mutations underlie primary congenital glaucoma with variable expressivity. J. Clin. Investig. 2016, 126, 2575–2587. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, S.; Kaur, K.; Rao, K.N.; Mandal, A.K.; Kaur, I.; Parikh, R.S.; Thomas, R. The Transcription Factor Gene FOXC1 Exhibits a Limited Role in Primary Congenital Glaucoma. Investig. Opthalmology Vis. Sci. 2009, 50, 75–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siggs, O.M.; Souzeau, E.; Taranath, D.A.; Dubowsky, A.; Chappell, A.; Zhou, T.; Javadiyan, S.; Nicholl, J.; Kearns, L.S.; Staffieri, S.E.; et al. Biallelic CPAMD8 Variants Are a Frequent Cause of Childhood and Juvenile Open-Angle Glaucoma. Ophthalmology 2020, 127, 758–766. [Google Scholar] [CrossRef]
- Bonet-Fernández, J.-M.; Aroca-Aguilar, J.-D.; Corton, M.; Ramírez, A.-I.; Alexandre-Moreno, S.; García-Antón, M.-T.; Salazar, J.-J.; Ferre-Fernández, J.-J.; Atienzar-Aroca, R.; Villaverde, C.; et al. CPAMD8 loss-of-function underlies non-dominant congenital glaucoma with variable anterior segment dysgenesis and abnormal extracellular matrix. Qual. Life Res. 2020, 139, 1209–1231. [Google Scholar] [CrossRef] [PubMed]
- Ferre-Fernández, J.-J.; Aroca-Aguilar, J.-D.; Medina-Trillo, C.; Bonet-Fernández, J.-M.; Méndez-Hernández, C.-D.; Morales-Fernández, L.; Corton, M.; Cabañero-Valera, M.-J.; Gut, M.; Tonda, R.; et al. Whole-Exome Sequencing of Congenital Glaucoma Patients Reveals Hypermorphic Variants in GPATCH3, a New Gene Involved in Ocular and Craniofacial Development. Sci. Rep. 2017, 7, 46175. [Google Scholar] [CrossRef] [Green Version]
- Morales-Cámara, S.; Alexandre-Moreno, S.; Bonet-Fernández, J.-M.; Atienzar-Aroca, R.; Aroca-Aguilar, J.-D.; Ferre-Fernández, J.-J.; Méndez, C.-D.; Morales, L.; Fernández-Sánchez, L.; Cuenca, N.; et al. Role of GUCA1C in Primary Congenital Glaucoma and in the Retina: Functional Evaluation in Zebrafish. Genes 2020, 11, 550. [Google Scholar] [CrossRef]
- Hollander, D.A.; Sarfarazi, M.; Stoilov, I.; Wood, I.S.; Fredrick, D.R.; Alvarado, J.A. Genotype and phenotype correlations in congenital glaucoma. Trans. Am. Ophthalmol. Soc. 2006, 104, 183–195. [Google Scholar]
- Alzuhairy, S.; Abu-Amero, K.K.; Al-Shahwan, S.; Edward, D.P. A Novel CYP1B1 Mutation with Congenital Glaucoma and Total Aniridia. Ophthalmic Genet. 2013, 36, 89–91. [Google Scholar] [CrossRef]
- Medina-Trillo, C.; Aroca-Aguilar, J.-D.; Ferre-Fernández, J.-J.; Alexandre-Moreno, S.; Morales, L.; Méndez-Hernández, C.-D.; García-Feijoo, J.; Escribano, J. Role of FOXC2 and PITX2 rare variants associated with mild functional alterations as modifier factors in congenital glaucoma. PLoS ONE 2019, 14, e0211029. [Google Scholar] [CrossRef]
- López-Garrido, M.-P.; Blanco-Marchite, C.; Sánchez-Sánchez, F.; Lóez-Sánchez, E.; Chaqués-Alepuz, V.; Campos-Mollo, E.; Salinas-Sánchez, A.; Escribano, J. Functional analysis of CYP1B1 mutations and association of heterozygous hypomorphic alleles with primary open-angle glaucoma. Clin. Genet. 2009, 77, 70–78. [Google Scholar] [CrossRef]
- Libby, R.T.; Smith, R.S.; Savinova, O.V.; Zabaleta, A.; Martin, J.E.; Gonzalez, F.J.; John, S.W.M. Modification of Ocular Defects in Mouse Developmental Glaucoma Models by Tyrosinase. Science 2003, 299, 1578–1581. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, S.; Sorenson, C.M.; Teixeira, L.; Dubielzig, R.R.; Peters, D.M.; Conway, S.J.; Jefcoate, C.R.; Sheibani, N. Cyp1b1 Mediates Periostin Regulation of Trabecular Meshwork Development by Suppression of Oxidative Stress. Mol. Cell. Biol. 2013, 33, 4225–4240. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, L.B.C.; Zhao, Y.; Dubielzig, R.R.; Sorenson, C.M.; Sheibani, N. Ultrastructural Abnormalities of the Trabecular Meshwork Extracellular Matrix in Cyp1b1-Deficient Mice. Veter. Pathol. 2015, 52, 397–403. [Google Scholar] [CrossRef] [Green Version]
- Pingili, A.K.; Jennings, B.L.; Mukherjee, K.; Akroush, W.; Gonzalez, F.J.; Malik, K.U. 6β-Hydroxytestosterone, a metabolite of testosterone generated by CYP1B1, contributes to vascular changes in angiotensin II-induced hypertension in male mice. Biol. Sex Differ. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Falero-Perez, J.; Larsen, M.C.; Teixeira, L.B.C.; Zhang, H.F.; Lindner, V.; Sorenson, C.M.; Jefcoate, C.R.; Sheibani, N. Targeted deletion of Cyp1b1 in pericytes results in attenuation of retinal neovascularization and trabecular meshwork dysgenesis. Trends Dev. Biol. 2019, 12, 1–12. [Google Scholar] [PubMed]
- Falero-Perez, J.; Sorenson, C.M.; Sheibani, N. Retinal astrocytes transcriptome reveals Cyp1b1 regulates the expression of genes involved in cell adhesion and migration. PLoS ONE 2020, 15, e0231752. [Google Scholar] [CrossRef] [PubMed]
- Malaplate-Armand, C. Astroglial CYP1B1 up-regulation in inflammatory/oxidative toxic conditions: IL-1β effect and protection by N-acetylcysteine. Toxicol. Lett. 2003, 138, 243–251. [Google Scholar] [CrossRef]
- Veith, A.C.; Aram, B.B.; Jiang, W.; Wang, L.; Zhou, G.; Jefcoate, C.R.; Couroucli, X.I.; Lingappan, K.; Moorthy, B. Mice Lacking the Cytochrome P450 1B1 Gene Are Less Susceptible to Hyperoxic Lung Injury Than Wild Type. Toxicol. Sci. 2018, 165, 462–474. [Google Scholar] [CrossRef]
- Falero-Perez, J.; Sorenson, C.M.; Sheibani, N. Cyp1b1-deficient retinal astrocytes are more proliferative and migratory and are protected from oxidative stress and inflammation. Am. J. Physiol. Physiol. 2019, 316, C767–C781. [Google Scholar] [CrossRef]
- Larsen, M.C.; Bushkofsky, J.R.; Gorman, T.; Adhami, V.; Mukhtar, H.; Wang, S.; Reeder, S.; Sheibani, N.; Jefcoate, C.R. Cytochrome P450 1B1: An unexpected modulator of liver fatty acid homeostasis. Arch. Biochem. Biophys. 2015, 571, 21–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Divanovic, S.; Dalli, J.; Jorge-Nebert, L.F.; Flick, L.M.; Gálvez-Peralta, M.; Boespflug, N.D.; Stankiewicz, T.E.; Fitzgerald, J.M.; Somarathna, M.; Karp, C.L.; et al. Contributions of the Three CYP1 Monooxygenases to Pro-Inflammatory and Inflammation-Resolution Lipid Mediator Pathways. J. Immunol. 2013, 191, 3347–3357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bushkofsky, J.R.; Maguire, M.; Larsen, M.C.; Foong, Y.H.; Jefcoate, C.R. Cyp1b1 affects external control of mouse hepatocytes, fatty acid homeostasis and signaling involving HNF4α and PPARα. Arch. Biochem. Biophys. 2016, 597, 30–47. [Google Scholar] [CrossRef] [Green Version]
- Timme-Laragy, A.R.; Noyes, P.D.; Buhler, D.R.; Di Giulio, R.T. CYP1B1 knockdown does not alter synergistic developmental toxicity of polycyclic aromatic hydrocarbons in zebrafish (Danio rerio). Mar. Environ. Res. 2008, 66, 85–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, A.L.; Eason, J.; Chawla, B.; Bohnsack, B.L. Cyp1b1 Regulates Ocular Fissure Closure Through a Retinoic Acid–Independent Pathway. Investig. Opthalmology Vis. Sci. 2017, 58, 1084–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massarsky, A.; Bone, A.J.; Dong, W.; Hinton, D.E.; Prasad, G.; Di Giulio, R.T. AHR2 morpholino knockdown reduces the toxicity of total particulate matter to zebrafish embryos. Toxicol. Appl. Pharmacol. 2016, 309, 63–76. [Google Scholar] [CrossRef]
- Williams, A.L.; Bohnsack, B.L. Neural crest derivatives in ocular development: Discerning the eye of the storm. Birth Defects Res. Part C Embryo Today Rev. 2015, 105, 87–95. [Google Scholar] [CrossRef] [Green Version]
- Brogna, S.; Wen, J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009, 16, 107–113. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Kague, E.; Gallagher, M.; Burke, S.; Parsons, M.; Franz-Odendaal, T.; Fisher, S. Skeletogenic Fate of Zebrafish Cranial and Trunk Neural Crest. PLoS ONE 2012, 7, e47394. [Google Scholar] [CrossRef]
- Chambers, D.; Wilson, L.; Maden, M.; Lumsden, A. RALDH-independent generation of retinoic acid during vertebrate embryogenesis by CYP1B1. Development 2007, 134, 1369–1383. [Google Scholar] [CrossRef] [Green Version]
- El-Brolosy, M.A.; Kontarakis, Z.; Rossi, A.; Kuenne, C.; Günther, S.; Fukuda, N.; Kikhi, K.; Boezio, G.L.M.; Takacs, C.M.; Lai, S.-L.; et al. Genetic compensation triggered by mutant mRNA degradation. Nat. Cell Biol. 2019, 568, 193–197. [Google Scholar] [CrossRef]
- Cavodeassi, F.; Wilson, S.W. Looking to the future of zebrafish as a model to understand the genetic basis of eye disease. Qual. Life Res. 2019, 138, 993–1000. [Google Scholar] [CrossRef] [Green Version]
- Rossi, A.; Kontarakis, Z.; Gerri, C.; Nolte, H.; Hölper, S.; Krüger, M.; Stainier, D. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nat. Cell Biol. 2015, 524, 230–233. [Google Scholar] [CrossRef]
- Ma, Z.P.; Chen, J. Nonsense mutations and genetic compensation response. Hereditas (Beijing) 2019, 41, 359–364. [Google Scholar]
- Morales-Fernandez, L.; De La Casa, J.M.M.; Garcia-Bella, J.; Mendez, C.; Saenz-Frances, F.; García, C.M.; Escribano, J.; Garcia-Feijoo, J. Clinical Variability of Primary Congenital Glaucoma in a Spanish Family with CYP1B1 Gene Mutations. J. Glaucoma 2015, 24, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Bejjani, B.A.; Stockton, D.W.; Lewis, R.A.; Tomey, K.F.; Dueker, D.K.; Jabak, M.; Astle, W.F.; Lupski, J.R. Multiple CYP1B1 mutations and incomplete penetrance in an inbred population segregating primary congenital glaucoma suggest frequent de novo events and a dominant modifier locus. Hum. Mol. Genet. 2000, 9, 367–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bidinost, C.; Hernandez, N.; Edward, D.P.; Al-Rajhi, A.; Lewis, R.A.; Lupski, J.R.; Stockton, D.W.; Bejjani, B.A. Of Mice and Men: Tyrosinase Modification of Congenital Glaucoma in Mice but Not in Humans. Investig. Opthalmology Vis. Sci. 2006, 47, 1486–1490. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.; Foernzler, D.; Loftus, S.K.; Liu, S.; McPherson, J.; Jungers, K.A.; Apte, S.S.; Pavan, W.J.; Beier, D.R. A defect in a novel ADAMTS family member is the cause of the belted white-spotting mutation. Development 2003, 130, 4665–4672. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, M.H.; Alrowaished, S.S.; Goody, M.F.; Crawford, B.D.; Henry, C.A. Laminin and Matrix metalloproteinase 11 regulate Fibronectin levels in the zebrafish myotendinous junction. Skelet. Muscle 2016, 6, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Talas, U.; Dunlop, J.; Khalaf, S.; Leigh, I.M.; Kelsell, D. Human Elastase 1: Evidence for Expression in the Skin and the Identification of a Frequent Frameshift Polymorphism. J. Investig. Dermatol. 2000, 114, 165–170. [Google Scholar] [CrossRef] [Green Version]
- Yagi, T.; Takeichi, M. Cadherin superfamily genes: Functions, genomic organization, and neurologic diversity. Genes Dev. 2000, 14, 1169–1180. [Google Scholar]
- Takeichi, M. Morphogenetic roles of classic cadherins. Curr. Opin. Cell Biol. 1995, 7, 619–627. [Google Scholar] [CrossRef]
- Norris, R.A.; Damon, B.; Mironov, V.; Kasyanov, V.; Ramamurthi, A.; Moreno-Rodriguez, R.; Trusk, T.; Potts, J.D.; Goodwin, R.L.; Davis, J.; et al. Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J. Cell. Biochem. 2007, 101, 695–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goonesinghe, A.; Luan, X.-M.; Hurlstone, A.; Garrod, D. Desmosomal cadherins in zebrafish epiboly and gastrulation. BMC Dev. Biol. 2012, 12, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtin, E.; Hickey, G.; Kamel, G.; Davidson, A.J.; Liao, E.C. Zebrafish wnt9a is expressed in pharyngeal ectoderm and is required for palate and lower jaw development. Mech. Dev. 2011, 128, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Dworkin, S.; Simkin, J.; Darido, C.; Partridge, D.D.; Georgy, S.R.; Caddy, J.; Wilanowski, T.; Lieschke, G.J.; Doggett, K.; Heath, J.; et al. Grainyhead-like 3 regulation of endothelin-1 in the pharyngeal endoderm is critical for growth and development of the craniofacial skeleton. Mech. Dev. 2014, 133, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.; Trainor, P.; Trainor, P. Craniofacial malformations: Intrinsic vs extrinsic neural crest cell defects in Treacher Collins and 22q11 deletion syndromes. Clin. Genet. 2006, 69, 471–479. [Google Scholar] [CrossRef]
- Jenny, M.J.; Karchner, S.I.; Franks, D.G.; Woodin, B.R.; Stegeman, J.J.; Hahn, M.E. Distinct Roles of Two Zebrafish AHR Repressors (AHRRa and AHRRb) in Embryonic Development and Regulating the Response to 2,3,7,8-Tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 2009, 110, 426–441. [Google Scholar] [CrossRef] [Green Version]
- Eferl, R.; Hoebertz, A.; Schilling, A.F.; Rath, M.; Karreth, F.; Kenner, L.; Amling, M.; Wagner, E.F. The Fos-related antigen Fra-1 is an activator of bone matrix formation. EMBO J. 2004, 23, 2789–2799. [Google Scholar] [CrossRef]
- Vaira, S.; Johnson, T.; Hirbe, A.C.; Alhawagri, M.; Anwisye, I.; Sammut, B.; O’Neal, J.; Zou, W.; Weilbaecher, K.N.; Faccio, R.; et al. RelB is the NF- B subunit downstream of NIK responsible for osteoclast differentiation. Proc. Natl. Acad. Sci. USA 2008, 105, 3897–3902. [Google Scholar] [CrossRef] [Green Version]
- Rowbotham, S.E.; Illingworth, N.A.; Daly, A.K.; Veal, G.J.; Boddy, A.V. Role of UDP-Glucuronosyltransferase Isoforms in 13-cis Retinoic Acid Metabolism in Humans. Drug Metab. Dispos. 2010, 38, 1211–1217. [Google Scholar] [CrossRef] [Green Version]
- Duester, G.; Mic, F.A.; Molotkov, A. Cytosolic retinoid dehydrogenases govern ubiquitous metabolism of retinol to retinaldehyde followed by tissue-specific metabolism to retinoic acid. Chem. Interactions 2003, 143–144, 201–210. [Google Scholar] [CrossRef]
- Fares-Taie, L.; Gerber, S.; Chassaing, N.; Clayton-Smith, J.; Hanein, S.; Silva, E.; Serey, M.; Serre, V.; Gérard, X.; Baumann, C.; et al. ALDH1A3 Mutations Cause Recessive Anophthalmia and Microphthalmia. Am. J. Hum. Genet. 2013, 92, 265–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casey, J.; Kawaguchi, R.; Morrissey, M.; Sun, H.; McGettigan, P.; Nielsen, J.E.; Conroy, J.; Regan, R.; Kenny, E.; Cormican, P.; et al. First implication of STRA6 mutations in isolated anophthalmia, microphthalmia, and coloboma: A new dimension to the STRA6 phenotype. Hum. Mutat. 2011, 32, 1417–1426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golzio, C.; Martinovic-Bouriel, J.; Thomas, S.; Mougou-Zrelli, S.; Grattagliano-Bessières, B.; Bonnière, M.; Delahaye, S.; Munnich, A.; Encha-Razavi, F.; Lyonnet, S.; et al. Matthew-Wood Syndrome Is Caused by Truncating Mutations in the Retinol-Binding Protein Receptor Gene STRA6. Am. J. Hum. Genet. 2007, 80, 1179–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roos, L.; Fang, M.; Dali, C.; Jensen, H.; Christoffersen, N.; Wu, B.; Zhang, J.; Xu, R.; Harris, P.; Xu, X.; et al. A homozygous mutation in a consanguineous family consolidates the role ofALDH1A3in autosomal recessive microphthalmia. Clin. Genet. 2013, 86, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Chawla, B.; Swain, W.; Williams, A.L.; Bohnsack, B.L. Retinoic Acid Maintains Function of Neural Crest–Derived Ocular and Craniofacial Structures in Adult Zebrafish. Investig. Opthalmology Vis. Sci. 2018, 59, 1924–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasuga, H.; Hosogane, N.; Matsuoka, K.; Mori, I.; Sakura, Y.; Shimakawa, K.; Shinki, T.; Suda, T.; Taketomi, S. Characterization of transgenic rats constitutively expressing vitamin D-24-hydroxylase gene. Biochem. Biophys. Res. Commun. 2002, 297, 1332–1338. [Google Scholar] [CrossRef]
- Grygiel-Górniak, B. Peroxisome proliferator-activated receptors and their ligands: Nutritional and clinical implications—A review. Nutr. J. 2014, 13, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhalla, S.; Ozalp, C.; Fang, S.; Xiang, L.; Kemper, J.K. Ligand-activated Pregnane X Receptor Interferes with HNF-4 Signaling by Targeting a Common Coactivator PGC-1α. J. Biol. Chem. 2004, 279, 45139–45147. [Google Scholar] [CrossRef] [Green Version]
- Maguire, M.; Larsen, M.C.; Vezina, C.M.; Quadro, L.; Kim, Y.-K.; Tanumihardjo, S.A.; Jefcoate, C.R. Cyp1b1 directs Srebp-mediated cholesterol and retinoid synthesis in perinatal liver; Association with retinoic acid activity during fetal development. PLoS ONE 2020, 15, e0228436. [Google Scholar] [CrossRef] [PubMed]
- Šmerdová, L.; Svobodová, J.; Kabátková, M.; Kohoutek, J.; Blažek, D.; Machala, M.; Vondráček, J. Upregulation of CYP1B1 expression by inflammatory cytokines is mediated by the p38 MAP kinase signal transduction pathway. Carcinogenesis 2014, 35, 2534–2543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]








| KEGG Term | Count | p | Genes |
|---|---|---|---|
| Steroid hormone biosynthesis | 7 | <0.0001 | hsd11b2, cyp11c1, srd5a2b, cyp1b1, ugt2a2, ugt1ab, cyp7a1 |
| PPAR signaling pathway | 6 | 0.006 | cpt2, ubb, aqp7, cyp7a1, pltp, cyp8b1 |
| Drug metabolism—other enzymes | 4 | 0.023 | zgc:103601, ugt2a2, tk1, ugt1ab |
| Retinol metabolism | 4 | 0.028 | ugt2a2, si:ch1073-13h15.3, ugt1ab, zgc:109982 |
| Metabolism of xenobiotics by cytochrome P450 | 4 | 0.028 | gstt2, cyp1b1, ugt2a2, ugt1ab |
| Primary bile acid biosynthesis | 3 | 0.049 | ch25h, cyp7a1, cyp8b1 |
| Steroid biosynthesis | 3 | 0.049 | cyp24a1, sc5d, cel.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexandre-Moreno, S.; Bonet-Fernández, J.-M.; Atienzar-Aroca, R.; Aroca-Aguilar, J.-D.; Escribano, J. Null cyp1b1 Activity in Zebrafish Leads to Variable Craniofacial Defects Associated with Altered Expression of Extracellular Matrix and Lipid Metabolism Genes. Int. J. Mol. Sci. 2021, 22, 6430. https://doi.org/10.3390/ijms22126430
Alexandre-Moreno S, Bonet-Fernández J-M, Atienzar-Aroca R, Aroca-Aguilar J-D, Escribano J. Null cyp1b1 Activity in Zebrafish Leads to Variable Craniofacial Defects Associated with Altered Expression of Extracellular Matrix and Lipid Metabolism Genes. International Journal of Molecular Sciences. 2021; 22(12):6430. https://doi.org/10.3390/ijms22126430
Chicago/Turabian StyleAlexandre-Moreno, Susana, Juan-Manuel Bonet-Fernández, Raquel Atienzar-Aroca, José-Daniel Aroca-Aguilar, and Julio Escribano. 2021. "Null cyp1b1 Activity in Zebrafish Leads to Variable Craniofacial Defects Associated with Altered Expression of Extracellular Matrix and Lipid Metabolism Genes" International Journal of Molecular Sciences 22, no. 12: 6430. https://doi.org/10.3390/ijms22126430
APA StyleAlexandre-Moreno, S., Bonet-Fernández, J. -M., Atienzar-Aroca, R., Aroca-Aguilar, J. -D., & Escribano, J. (2021). Null cyp1b1 Activity in Zebrafish Leads to Variable Craniofacial Defects Associated with Altered Expression of Extracellular Matrix and Lipid Metabolism Genes. International Journal of Molecular Sciences, 22(12), 6430. https://doi.org/10.3390/ijms22126430