PRP4 Promotes Skin Cancer by Inhibiting Production of Melanin, Blocking Influx of Extracellular Calcium, and Remodeling Cell Actin Cytoskeleton
Abstract
:1. Introduction
2. Results
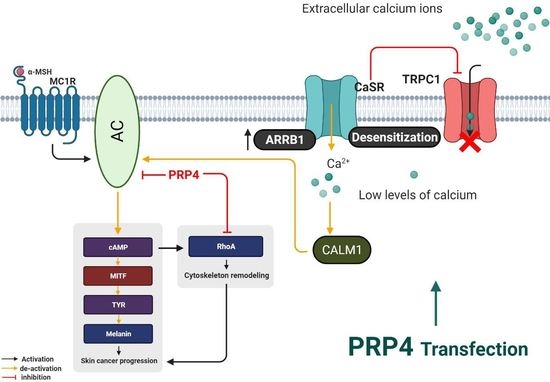
2.1. PRP4 Inhibits the Production of Melanin in B16F10 Cells and Promotes Skin Cancer
2.2. PRP4 Regulates ARRB1-CaSR Pathway
2.3. PRP4 Overexpression Reduces the Influx of Ca2+ and Downregulates Calmodulin
2.4. PRP4-Regulated CaSR-Induced Ca2+ Influx Occurs through the TRPC1 Channel in B16F10 Cells
2.5. PRP4 Changes the Morphology of B16F10 by Regulating the AC–cAMP–RhoA Pathway
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture and Double Transient Transfections
4.3. Plasmid Transfection and Gene Knock Down by siRNA
4.4. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
4.5. Relative qRT-PCR Analysis
4.6. F-Actin Staining
4.7. Cal-520 AM Assay
4.8. cAMP Assay
4.9. Animal Study Protocol
4.10. Measurement of Melanin Content
4.11. Western Blot
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PRP4 | Pre-mRNA processing factor 4B |
| EMT | Epithelial-mesenchymal transition |
| AC | Adenylyl cyclase |
| CaSR | Calcium-sensing receptor |
| RhoA | Ras homolog family member A |
| TRP-1 | Tyrosinase-related protein |
| MITF | Microphthalmia-associated transcription factor |
| CREB | cAMP response element-binding protein |
| ERK | Extracellular-regulated kinase |
| α-MSH | Alpha-melanocyte stimulating hormone |
| GPCR | G-protein-coupled receptor |
| GRKs | G protein-coupled receptor kinase |
| TRP | Transient receptor potential |
| TRPC1 | Transient receptor potential canonical 1 |
| cAMP | Cyclic adenosine 3′,5′-monophosphate |
| MC1R | Melanocortin 1 receptor |
References
- Martínez-Esparza, M.; Jiménez-Cervantes, C.; Beermann, F.; Aparicio, P.; Lozano, J.A.; García-Borrón, J.C. Transforming growth factor-beta1 inhibits basal melanogenesis in B16/F10 mouse melanoma cells by increasing the rate of degradation of tyrosinase and tyrosinase-related protein-1. J. Biol. Chem. 1997, 272, 3967–3972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poste, G.; Doll, J.; Fidler, I.J. Interactions among clonal subpopulations affect stability of the metastatic phenotype in polyclonal populations of B16 melanoma cells. Proc. Natl. Acad. Sci. USA 1981, 78, 6226–6230. [Google Scholar] [CrossRef] [Green Version]
- Calliste, C.A.; Trouillas, P.; Allais, D.P.; Simon, A.; Duroux, J.L. Free radical scavenging activities measured by electron spin resonance spectroscopy and B16 cell antiproliferative behaviors of seven plants. J. Agric. Food Chem. 2001, 49, 3321–3327. [Google Scholar] [CrossRef]
- Jin, K.S.; Oh, Y.N.; Hyun, S.K.; Kwon, H.J.; Kim, B.W. Betulinic acid isolated from Vitis amurensis root inhibits 3-isobutyl-1-methylxanthine induced melanogenesis via the regulation of MEK/ERK and PI3K/Akt pathways in B16F10 cells. Food Chem. Toxicol. 2014, 68, 38–43. [Google Scholar] [CrossRef]
- Bentley, N.J.; Eisen, T.; Goding, C.R. Melanocyte-specific expression of the human tyrosinase promoter: Activation by the microphthalmia gene product and role of the initiator. Mol. Cell. Biol. 1994, 14, 7996–8006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertolotto, C.; Buscà, R.; Abbe, P.; Bille, K.; Aberdam, E.; Ortonne, J.P.; Ballotti, R. Different cis-acting elements are involved in the regulation of TRP1 and TRP2 promoter activities by cyclic AMP: Pivotal role of M boxes (GTCATGTGCT) and of microphthalmia. Mol. Cell. Biol. 1998, 18, 694–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.C.; Chang, S.J.; Wu, C.Y.; Ke, H.J.; Chang, T.M. [6]-Shogaol inhibits α-MSH-induced melanogenesis through the acceleration of ERK and PI3K/Akt-mediated MITF degradation. BioMed Res. Int. 2014, 2014, 842569. [Google Scholar] [CrossRef] [Green Version]
- Gilchrest, B.A.; Eller, M.S.; Geller, A.C.; Yaar, M. The pathogenesis of melanoma induced by ultraviolet radiation. N. Engl. J. Med. 1999, 340, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Halder, R.M.; Bang, K.M. Skin cancer in blacks in the United States. Dermatol. Clin. 1988, 6, 397–405. [Google Scholar] [CrossRef]
- Kaidbey, K.H.; Agin, P.P.; Sayre, R.M.; Kligman, A.M. Photoprotection by melanin—A comparison of black and Caucasian skin. J. Am. Acad. Dermatol. 1979, 1, 249–260. [Google Scholar] [CrossRef]
- Szabó, G.; Gerald, A.B.; Pathak, M.A.; Fitzpatrick, T.B. Racial differences in the fate of melanosomes in human epidermis. Nature 1969, 222, 1081–1082. [Google Scholar] [CrossRef]
- Kobayashi, N.; Nakagawa, A.; Muramatsu, T.; Yamashina, Y.; Shirai, T.; Hashimoto, M.W.; Ishigaki, Y.; Ohnishi, T.; Mori, T. Supranuclear melanin caps reduce ultraviolet induced DNA photoproducts in human epidermis. J. Investig. Dermatol. 1998, 110, 806–810. [Google Scholar] [CrossRef] [Green Version]
- Vahe, C.; Benomar, K.; Espiard, S.; Coppin, L.; Jannin, A.; Odou, M.F.; Vantyghem, M.C. Diseases associated with calcium-sensing receptor. Orphanet J. Rare Dis. 2017, 12, 19. [Google Scholar] [CrossRef] [Green Version]
- Thakker, R.V. Calcium-sensing receptor: Role in health and disease. Indian J. Endocrinol. Metab. 2012, 16 (Suppl. 2), S213–S216. [Google Scholar]
- Lorenz, S.; Frenzel, R.; Paschke, R.; Breitwieser, G.E.; Miedlich, S.U. Functional desensitization of the extracellular calcium-sensing receptor is regulated via distinct mechanisms: Role of G protein-coupled receptor kinases, protein kinase C and beta-arrestins. Endocrinology 2007, 148, 2398–2404. [Google Scholar] [CrossRef] [Green Version]
- Pi, M.; Oakley, R.H.; Gesty-Palmer, D.; Cruickshank, R.D.; Spurney, R.F.; Luttrell, L.M.; Quarles, L.D. Beta-arrestin- and G protein receptor kinase-mediated calcium-sensing receptor desensitization. Mol. Endocrinol. 2005, 19, 1078–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes-Ibarra, A.P.; García-Regalado, A.; Ramírez-Rangel, I.; Esparza-Silva, A.L.; Valadez-Sánchez, M.; Vázquez-Prado, J.; Reyes-Cruz, G. Calcium-sensing receptor endocytosis links extracellular calcium signaling to parathyroid hormone-related peptide secretion via a Rab11a-dependent and AMSH-sensitive mechanism. Mol. Endocrinol. 2007, 21, 1394–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nesbit, M.A.; Hannan, F.M.; Howles, S.A.; Reed, A.A.; Cranston, T.; Thakker, C.E.; Gregory, L.; Rimmer, A.J.; Rust, N.; Graham, U.; et al. Mutations in AP2S1 cause familial hypocalciuric hypercalcemia type 3. Nat. Genet. 2013, 45, 93–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, X.; Northup, J.K.; Ray, K. Large putative PEST-like sequence motif at the carboxyl tail of human calcium receptor directs lysosomal degradation and regulates cell surface receptor level. J. Biol. Chem. 2012, 287, 4165–4176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanyaloglu, A.C.; von Zastrow, M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 537–568. [Google Scholar] [CrossRef] [Green Version]
- Moore, C.A.; Milano, S.K.; Benovic, J.L. Regulation of receptor trafficking by GRKs and arrestins. Annu. Rev. Physiol. 2007, 69, 451–482. [Google Scholar] [CrossRef] [PubMed]
- DeWire, S.M.; Ahn, S.; Lefkowitz, R.J.; Shenoy, S.K. Beta-arrestins and cell signaling. Annu. Rev. Physiol. 2007, 69, 483–510. [Google Scholar] [CrossRef] [Green Version]
- Ambudkar, I.S.; Ong, H.L.; Liu, X.; Bandyopadhyay, B.C.; Cheng, K.T. TRPC1: The link between functionally distinct store-operated calcium channels. Cell Calcium 2007, 42, 213–223. [Google Scholar] [CrossRef] [PubMed]
- El Hiani, Y.; Ahidouch, A.; Lehen’kyi, V.Y.; Hague, F.; Gouilleux, F.; Mentaverri, R.; Kamel, S.; Lassoued, K.; Brûlé, G.; Ouadid-Ahidouch, H. Extracellular signal-regulated kinases 1 and 2 and TRPC1 channels are required for calcium-sensing receptor-stimulated MCF-7 breast cancer cell proliferation. Cell. Physiol. Biochem. 2009, 23, 335–346. [Google Scholar] [CrossRef]
- Tiruppathi, C.; Ahmmed, G.U.; Vogel, S.M.; Malik, A.B. Ca2+ signaling, TRP channels, and endothelial permeability. Microcirculation 2006, 13, 693–708. [Google Scholar] [CrossRef]
- Howe, A.K. Regulation of actin-based cell migration by cAMP/PKA. Biochim. Biophys. Acta 2004, 1692, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Howe, A.K. Cross-talk between calcium and protein kinase A in the regulation of cell migration. Curr. Opin. Cell Biol. 2011, 23, 554–561. [Google Scholar] [CrossRef] [Green Version]
- Whittard, J.D.; Akiyama, S.K. Positive regulation of cell-cell and cell-substrate adhesion by protein kinase A. J. Cell Sci. 2001, 114 (Pt 18), 3265–3272. [Google Scholar] [CrossRef]
- Plopper, G.E.; Huff, J.L.; Rust, W.L.; Schwartz, M.A.; Quaranta, V. Antibody-induced activation of beta1 integrin receptors stimulates cAMP-dependent migration of breast cells on laminin-5. Mol. cell Biol. Res. Commun. 2000, 4, 129–135. [Google Scholar] [CrossRef]
- Grieshaber, N.A.; Boitano, S.; Ji, I.; Mather, J.P.; Ji, T.H. Differentiation of granulosa cell line: Follicle-stimulating hormone induces formation of lamellipodia and filopodia via the adenylyl cyclase/cyclic adenosine monophosphate signal. Endocrinology 2000, 141, 3461–3470. [Google Scholar] [CrossRef]
- Feoktistov, I.; Goldstein, A.E.; Biaggioni, I. Cyclic AMP and protein kinase A stimulate Cdc42: Role of A(2) adenosine receptors in human mast cells. Mol. Pharmacol. 2000, 58, 903–910. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, K.L.; Mercurio, A.M. Protein kinase A regulates Rac and is required for the growth factor-stimulated migration of carcinoma cells. J. Biol. Chem. 2001, 276, 47895–47900. [Google Scholar] [CrossRef] [Green Version]
- Dunlap, M.K.; Donaldson, D.J. Effect of cAMP and related compounds on newt epidermal cell migration both in vivo and in vitro. J. Exp. Zool. 1980, 212, 13–19. [Google Scholar] [CrossRef]
- Smirnov, V.N.; Antonov, A.S.; Antonova, G.N.; Romanov, Y.A.; Kabaeva, N.V.; Tchertikhina, I.V.; Lukashev, M.E. Effects of forskolin and phorbol-myristate-acetate on cytoskeleton, extracellular matrix and protein phosphorylation in human endothelial cells. J. Mol. Cell. Cardiol. 1989, 21 (Suppl. 1), 3–11. [Google Scholar] [CrossRef]
- Goldman, J.E.; Abramson, B. Cyclic AMP-induced shape changes of astrocytes are accompanied by rapid depolymerization of actin. Brain Res. 1990, 528, 189–196. [Google Scholar] [CrossRef]
- Rosenberg, G.H.; Alahari, S.K.; Käufer, N.F. prp4 from Schizosaccharomyces pombe, a mutant deficient in pre-mRNA splicing isolated using genes containing artificial introns. Mol. Gen. Genet. MGG 1991, 226, 305–309. [Google Scholar] [CrossRef]
- Kojima, T.; Zama, T.; Wada, K.; Onogi, H.; Hagiwara, M. Cloning of human PRP4 reveals interaction with Clk1. J. Biol. Chem. 2001, 276, 32247–32256. [Google Scholar] [CrossRef] [Green Version]
- Lützelberger, M.; Käufer, N.F. The Prp4 kinase: Its substrates, function and regulation in pre-mRNA splicing. In Protein Phosphorylation in Human Health; InTech: London, UK, 2012. [Google Scholar]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Schwelnus, W.; Richert, K.; Opitz, F.; Groß, T.; Habara, Y.; Tani, T.; Käufer, N.F. Fission yeast Prp4p kinase regulates pre-mRNA splicing by phosphorylating a non-SR-splicing factor. EMBO Rep. 2001, 2, 35–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montembault, E.; Dutertre, S.; Prigent, C.; Giet, R. PRP4 is a spindle assembly checkpoint protein required for MPS1, MAD1, and MAD2 localization to the kinetochores. J. Cell Biol. 2007, 179, 601–609. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.; Ahn, Y.-T.; McPherson, L.; Clayberger, C.; Krensky, A.M. Interaction of PRP4 with Krüppel-like factor 13 regulates CCL5 transcription. J. Immunol. 2007, 178, 7081–7087. [Google Scholar] [CrossRef]
- Gao, Q.; Mechin, I.; Kothari, N.; Guo, Z.; Deng, G.; Haas, K.; McManus, J.; Hoffmann, D.; Wang, A.; Wiederschain, D. Evaluation of cancer dependence and druggability of PRP4 kinase using cellular, biochemical, and structural approaches. J. Biol. Chem. 2013, 288, 30125–30138. [Google Scholar] [CrossRef] [Green Version]
- Islam, S.U.; Shehzad, A.; Sonn, J.K.; Lee, Y.S. PRPF overexpression induces drug resistance through actin cytoskeleton rearrangement and epithelial-mesenchymal transition. Oncotarget 2017, 8, 56659. [Google Scholar] [CrossRef] [Green Version]
- Islam, S.U.; Ahmed, M.B.; Lee, S.J.; Shehzad, A.; Sonn, J.K.; Kwon, O.S.; Lee, Y.S. PRP4 kinase induces actin rearrangement and epithelial-mesenchymal transition through modulation of the actin-binding protein cofilin. Exp. Cell Res. 2018, 369, 158–165. [Google Scholar] [CrossRef]
- Bae-Harboe, Y.S.; Park, H.Y. Tyrosinase: A central regulatory protein for cutaneous pigmentation. J. Investig. Dermatol. 2012, 132, 2678–2680. [Google Scholar] [CrossRef] [Green Version]
- D’Mello, S.A.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling Pathways in Melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef] [Green Version]
- Watson, E.L.; Jacobson, K.L.; Singh, J.C.; Idzerda, R.; Ott, S.M.; DiJulio, D.H.; Wong, S.T.; Storm, D.R. The type 8 adenylyl cyclase is critical for Ca2+ stimulation of cAMP accumulation in mouse parotid acini. J. Biol. Chem. 2000, 275, 14691–14699. [Google Scholar] [CrossRef] [Green Version]
- Oishi, A.; Makita, N.; Sato, J.; Iiri, T. Regulation of RhoA signaling by the cAMP-dependent phosphorylation of RhoGDIα. J. Biol. Chem. 2012, 287, 38705–38715. [Google Scholar] [CrossRef] [Green Version]
- Chung, S.; Lim, G.J.; Lee, J.Y. Quantitative analysis of melanin content in a three-dimensional melanoma cell culture. Sci. Rep. 2019, 9, 780. [Google Scholar] [CrossRef]
- Peters, A.A.; Simpson, P.T.; Bassett, J.J.; Lee, J.M.; Da Silva, L.; Reid, L.E.; Song, S.; Parat, M.O.; Lakhani, S.R.; Kenny, P.A.; et al. Calcium channel TRPV6 as a potential therapeutic target in estrogen receptor-negative breast cancer. Mol. Cancer Ther. 2012, 11, 2158–2168. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; He, Z.; Zhou, K.; Cheng, J.; Yao, H.; Lu, D.; Cai, R.; Jin, Y.; Dong, B.; Xu, Y.; et al. Essential role of TRPC6 channels in G2/M phase transition and development of human glioma. J. Natl. Cancer Inst. 2010, 102, 1052–1068. [Google Scholar] [CrossRef] [Green Version]
- Tsavaler, L.; Shapero, M.H.; Morkowski, S.; Laus, R. Trp-p8, a novel prostate-specific gene, is up-regulated in prostate cancer and other malignancies and shares high homology with transient receptor potential calcium channel proteins. Cancer Res. 2001, 61, 3760–3769. [Google Scholar]
- Stewart, T.A.; Yapa, K.T.; Monteith, G.R. Altered calcium signaling in cancer cells. Biochim. Biophys. Acta 2015, 1848 (10 Pt B), 2502–2511. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, M.B.; Islam, S.U.; Lee, Y.S. PRP4 Promotes Skin Cancer by Inhibiting Production of Melanin, Blocking Influx of Extracellular Calcium, and Remodeling Cell Actin Cytoskeleton. Int. J. Mol. Sci. 2021, 22, 6992. https://doi.org/10.3390/ijms22136992
Ahmed MB, Islam SU, Lee YS. PRP4 Promotes Skin Cancer by Inhibiting Production of Melanin, Blocking Influx of Extracellular Calcium, and Remodeling Cell Actin Cytoskeleton. International Journal of Molecular Sciences. 2021; 22(13):6992. https://doi.org/10.3390/ijms22136992
Chicago/Turabian StyleAhmed, Muhammad Bilal, Salman Ul Islam, and Young Sup Lee. 2021. "PRP4 Promotes Skin Cancer by Inhibiting Production of Melanin, Blocking Influx of Extracellular Calcium, and Remodeling Cell Actin Cytoskeleton" International Journal of Molecular Sciences 22, no. 13: 6992. https://doi.org/10.3390/ijms22136992
APA StyleAhmed, M. B., Islam, S. U., & Lee, Y. S. (2021). PRP4 Promotes Skin Cancer by Inhibiting Production of Melanin, Blocking Influx of Extracellular Calcium, and Remodeling Cell Actin Cytoskeleton. International Journal of Molecular Sciences, 22(13), 6992. https://doi.org/10.3390/ijms22136992